20n/act is the data aggregation and prediction system for bioengineering. For a target molecule, 20n/act predicts DNA insertions into cells (usually a microbe such as E. coli or S. cerevisiae) that modify the cell. These modified cells make the target molecule by fermentation from sugar. We call these "target molecules/chemicals" the bioreachables. The system predicted/invented the first bio-route to Acetaminophen/Tylenol/APAP. Read more on our blog post. The technical details of the APAP work can be found in patents applications on coli and yeast fermentation.
See predicted DNA for 11 sample molecules at Bioreachables Preview (Login:Pass = public:preview). Due to limitations we can only make a preview version available. If you'd like the full version please contact us.
Checkout the repo. Follow instructions to run to create the database and prediction corpus. If you'd rather get a pre-packaged DB without creating it yourself please contact us. The codebase is public to further the state-of-the-art in automating biological engineering/synthetic biology. Some modules are specific to microbes, but most of the predictive stack deals with host-agnostic enzymatic biochemistry.
Answers "what DNA do I insert if I want to make my chemical?"
| Module | Function | Code | |
|---|---|---|---|
| 1 | Installer | Integrates heterogeneous raw data | Code:com.act.reachables.initdb Run:Instructions |
| 2 | Reaction operator (RO) inference | Mines rules of enzymatic catalysis | Code:biointerpretation module |
| 2 | Structure Activity Relationship (SAR) inference | Mines substrate specificities | Code:biointerpretation module |
| 3 | Biointerpretation | Mechanistic validation of enzymatic transforms (using ROs) | Code:com.act.biointerpretation.BiointerpretationDriver Run:Instructions |
| 4 | Reachables computation | Exhaustively enumerates all biosynthesizable chemicals | Code:com.act.reachables.reachables Code:com.act.reachables.postprocess_reachables Run:Instructions |
| 5 | Cascades computation | Exhaustively enumerates all enzymatic routes from metabolic natives to bioreachable target | Code:com.act.reachables.cascades Run:Instructions |
| 6 | DNA designer | Computes protein & DNA design (coli specific) for each non-natural enzymatic path | Code:org.twentyn.proteintodna.ProteinToDNADriver Run:Instructions |
| 7 | Application miner | Mines chemical applications using web searches [Bing] | Code:act.installer.bing.BingSearcher Run:Instructions |
| 8 | Enzymatic biochemistry NLP | Text -> Chemical tokens -> Biologically feasible reactions using ROs | Code:act.shared.TextToRxns Frontend:TextToRxnsUI |
| 9 | Patent search | Chemical -> Patents | Code:act.installer.reachablesexplorer.PatentFinder Run:Instructions |
| 10 | Bioreachables wiki | Aggregates reachables, cascades, use cases, protein and DNA designs into a user friendly wiki interface | Documentation |
Answers "Is my bio-engineered cell doing what I want it to?"
| Module | Function | Code | |
|---|---|---|---|
| 1 | LCMS: Untargeted metabolomics | Deep-learnt signal processing to identify all chemical [side]effects of DNA engineering on cell | Code:DeepLearningLcmsPeak Code:com.act.lcms.UntargetedMetabolomics |
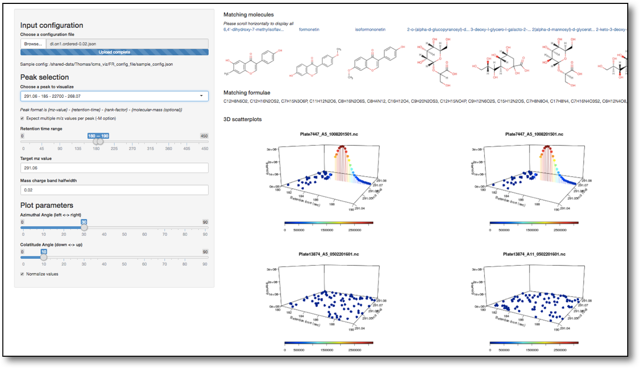
| 2 | LCMS: Comparative visualization | Visualizing traces side-by-side from untargeted evaluation of over and underexpressed peaks | Doc:LCMSDataVisualisation |
Answers "Can I use bio-production to make this chemical at scale?"
| Module | Function | Code | |
|---|---|---|---|
| 1 | Cost model: Manufacturing unit economics for large scale production | It backcalculates cell efficiency (yield, titers, productivity) objectives based on given COGS ($ per ton) of target chemical. From cell efficiency objectives it guesstimates the R&D investment (money and time) and ROI expectations | Code:act.installer.bing.CostModel Code (viz server):costModelUI Source model:XLS |
Code licensed under the GNU General Public License v3.0. If an alternative license is desired, please contact 20n.