diff --git a/ci/bundle_custom_data.py b/ci/bundle_custom_data.py
index 8d391437..cf56d530 100644
--- a/ci/bundle_custom_data.py
+++ b/ci/bundle_custom_data.py
@@ -39,6 +39,7 @@
"brats_mri_axial_slices_generative_diffusion",
"vista3d",
"maisi_ct_generative",
+ "vista2d",
]
# This list is used for our CI tests to determine whether a bundle needs to be tested after downloading
diff --git a/ci/unit_tests/test_vista2d.py b/ci/unit_tests/test_vista2d.py
new file mode 100644
index 00000000..0eb5fb5d
--- /dev/null
+++ b/ci/unit_tests/test_vista2d.py
@@ -0,0 +1,125 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import os
+import shutil
+import sys
+import tempfile
+import unittest
+
+import matplotlib.pyplot as plt
+import numpy as np
+from monai.bundle import create_workflow
+from parameterized import parameterized
+from utils import check_workflow
+
+TEST_CASE_TRAIN = [{"bundle_root": "models/vista2d", "mode": "train", "train#trainer#max_epochs": 1}]
+
+TEST_CASE_INFER = [{"bundle_root": "models/vista2d", "mode": "infer"}]
+
+
+def test_order(test_name1, test_name2):
+ def get_order(name):
+ if "train" in name:
+ return 1
+ if "infer" in name:
+ return 2
+ return 3
+
+ return get_order(test_name1) - get_order(test_name2)
+
+
+class TestVista2d(unittest.TestCase):
+ def setUp(self):
+ self.dataset_dir = tempfile.mkdtemp()
+ self.tmp_output_dir = os.path.join(self.dataset_dir, "output")
+ os.makedirs(self.tmp_output_dir, exist_ok=True)
+ self.dataset_size = 5
+ input_shape = (256, 256)
+ for s in range(self.dataset_size):
+ test_image = np.random.randint(low=0, high=2, size=input_shape).astype(np.int8)
+ test_label = np.random.randint(low=0, high=2, size=input_shape).astype(np.int8)
+ image_filename = os.path.join(self.dataset_dir, f"image_{s}.png")
+ label_filename = os.path.join(self.dataset_dir, f"label_{s}.png")
+ plt.imsave(image_filename, test_image, cmap="gray")
+ plt.imsave(label_filename, test_label, cmap="gray")
+
+ self.bundle_root = "models/vista2d"
+ sys.path = [self.bundle_root] + sys.path
+ from scripts.workflow import VistaCell
+
+ self.workflow = VistaCell
+
+ def tearDown(self):
+ shutil.rmtree(self.dataset_dir)
+
+ @parameterized.expand([TEST_CASE_INFER])
+ def test_infer_config(self, override):
+ # update override with dataset dir
+ override["dataset#data"] = [
+ {
+ "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
+ "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
+ }
+ for s in range(self.dataset_size)
+ ]
+ override["output_dir"] = self.tmp_output_dir
+ workflow = create_workflow(
+ workflow_name=self.workflow,
+ config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
+ meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
+ **override,
+ )
+
+ # check_properties=False, need to add monai service properties later
+ check_workflow(workflow, check_properties=False)
+
+ expected_output_file = os.path.join(self.tmp_output_dir, f"image_{self.dataset_size-1}.tif")
+ self.assertTrue(os.path.isfile(expected_output_file))
+
+ @parameterized.expand([TEST_CASE_TRAIN])
+ def test_train_config(self, override):
+ # update override with dataset dir
+ override["train#dataset#data"] = [
+ {
+ "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
+ "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
+ }
+ for s in range(self.dataset_size)
+ ]
+ override["dataset#data"] = override["train#dataset#data"]
+
+ workflow = create_workflow(
+ workflow_name=self.workflow,
+ config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
+ meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
+ **override,
+ )
+
+ # check_properties=False, need to add monai service properties later
+ check_workflow(workflow, check_properties=False)
+
+ # follow up to use trained weights and test eval
+ override["mode"] = "eval"
+ override["pretrained_ckpt_name"] = "model.pt"
+ workflow = create_workflow(
+ workflow_name=self.workflow,
+ config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
+ meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
+ **override,
+ )
+ check_workflow(workflow, check_properties=False)
+
+
+if __name__ == "__main__":
+ loader = unittest.TestLoader()
+ loader.sortTestMethodsUsing = test_order
+ unittest.main(testLoader=loader)
diff --git a/ci/unit_tests/test_vista2d_dist.py b/ci/unit_tests/test_vista2d_dist.py
new file mode 100644
index 00000000..b3cdf1c2
--- /dev/null
+++ b/ci/unit_tests/test_vista2d_dist.py
@@ -0,0 +1,70 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import os
+import shutil
+import sys
+import tempfile
+import unittest
+
+import matplotlib.pyplot as plt
+import numpy as np
+import torch
+from parameterized import parameterized
+from utils import export_config_and_run_mgpu_cmd
+
+TEST_CASE_TRAIN_MGPU = [{"bundle_root": "models/vista2d", "workflow_type": "train", "train#trainer#max_epochs": 2}]
+
+
+class TestVista2d(unittest.TestCase):
+ def setUp(self):
+ self.dataset_dir = tempfile.mkdtemp()
+ self.dataset_size = 5
+ input_shape = (256, 256)
+ for s in range(self.dataset_size):
+ test_image = np.random.randint(low=0, high=2, size=input_shape).astype(np.int8)
+ test_label = np.random.randint(low=0, high=2, size=input_shape).astype(np.int8)
+ image_filename = os.path.join(self.dataset_dir, f"image_{s}.png")
+ label_filename = os.path.join(self.dataset_dir, f"label_{s}.png")
+ plt.imsave(image_filename, test_image, cmap="gray")
+ plt.imsave(label_filename, test_label, cmap="gray")
+
+ self.bundle_root = "models/vista2d"
+ sys.path = [self.bundle_root] + sys.path
+
+ def tearDown(self):
+ shutil.rmtree(self.dataset_dir)
+
+ @parameterized.expand([TEST_CASE_TRAIN_MGPU])
+ def test_train_mgpu_config(self, override):
+ override["train#dataset#data"] = [
+ {
+ "image": os.path.join(self.dataset_dir, f"image_{s}.png"),
+ "label": os.path.join(self.dataset_dir, f"label_{s}.png"),
+ }
+ for s in range(self.dataset_size)
+ ]
+ override["dataset#data"] = override["train#dataset#data"]
+
+ output_path = os.path.join(self.bundle_root, "configs/train_override.json")
+ n_gpu = torch.cuda.device_count()
+ export_config_and_run_mgpu_cmd(
+ config_file=os.path.join(self.bundle_root, "configs/hyper_parameters.yaml"),
+ meta_file=os.path.join(self.bundle_root, "configs/metadata.json"),
+ custom_workflow="scripts.workflow.VistaCell",
+ override_dict=override,
+ output_path=output_path,
+ ngpu=n_gpu,
+ )
+
+
+if __name__ == "__main__":
+ unittest.main()
diff --git a/ci/unit_tests/utils.py b/ci/unit_tests/utils.py
index 7fda766d..eba94701 100644
--- a/ci/unit_tests/utils.py
+++ b/ci/unit_tests/utils.py
@@ -23,7 +23,7 @@ def export_overrided_config(config_file, override_dict, output_path):
ConfigParser.export_config_file(parser.config, output_path, indent=4)
-def produce_mgpu_cmd(config_file, meta_file, logging_file, nnodes=1, nproc_per_node=2):
+def produce_mgpu_cmd(config_file, meta_file, logging_file=None, nnodes=1, nproc_per_node=2):
cmd = [
"torchrun",
"--standalone",
@@ -34,20 +34,43 @@ def produce_mgpu_cmd(config_file, meta_file, logging_file, nnodes=1, nproc_per_n
"run",
"--config_file",
config_file,
- "--logging_file",
- logging_file,
"--meta_file",
meta_file,
]
+ if logging_file is not None:
+ cmd.extend(["--logging_file", logging_file])
+ return cmd
+
+
+def produce_custom_workflow_mgpu_cmd(
+ custom_workflow, config_file, meta_file, logging_file=None, nnodes=1, nproc_per_node=2
+):
+ cmd = [
+ "torchrun",
+ "--standalone",
+ f"--nnodes={nnodes}",
+ f"--nproc_per_node={nproc_per_node}",
+ "-m",
+ "monai.bundle",
+ "run_workflow",
+ custom_workflow,
+ "--config_file",
+ config_file,
+ "--meta_file",
+ meta_file,
+ ]
+ if logging_file is not None:
+ cmd.extend(["--logging_file", logging_file])
return cmd
def export_config_and_run_mgpu_cmd(
config_file,
meta_file,
- logging_file,
override_dict,
output_path,
+ custom_workflow=None,
+ logging_file=None,
workflow_type="train",
nnode=1,
ngpu=2,
@@ -68,9 +91,19 @@ def export_config_and_run_mgpu_cmd(
check_result = engine.check_properties()
if check_result is not None and len(check_result) > 0:
raise ValueError(f"check properties for overrided mgpu configs failed: {check_result}")
- cmd = produce_mgpu_cmd(
- config_file=output_path, meta_file=meta_file, logging_file=logging_file, nnodes=nnode, nproc_per_node=ngpu
- )
+ if custom_workflow is None:
+ cmd = produce_mgpu_cmd(
+ config_file=output_path, meta_file=meta_file, logging_file=logging_file, nnodes=nnode, nproc_per_node=ngpu
+ )
+ else:
+ cmd = produce_custom_workflow_mgpu_cmd(
+ custom_workflow=custom_workflow,
+ config_file=output_path,
+ meta_file=meta_file,
+ logging_file=logging_file,
+ nnodes=nnode,
+ nproc_per_node=ngpu,
+ )
env = os.environ.copy()
# ensure customized library can be loaded in subprocess
env["PYTHONPATH"] = override_dict.get("bundle_root", ".")
diff --git a/models/vista2d/LICENSE b/models/vista2d/LICENSE
new file mode 100644
index 00000000..bdd91f8f
--- /dev/null
+++ b/models/vista2d/LICENSE
@@ -0,0 +1,649 @@
+Code License
+
+This license applies to all files except the model weights in the directory.
+
+ Apache License
+ Version 2.0, January 2004
+ http://www.apache.org/licenses/
+
+ TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
+
+ 1. Definitions.
+
+ "License" shall mean the terms and conditions for use, reproduction,
+ and distribution as defined by Sections 1 through 9 of this document.
+
+ "Licensor" shall mean the copyright owner or entity authorized by
+ the copyright owner that is granting the License.
+
+ "Legal Entity" shall mean the union of the acting entity and all
+ other entities that control, are controlled by, or are under common
+ control with that entity. For the purposes of this definition,
+ "control" means (i) the power, direct or indirect, to cause the
+ direction or management of such entity, whether by contract or
+ otherwise, or (ii) ownership of fifty percent (50%) or more of the
+ outstanding shares, or (iii) beneficial ownership of such entity.
+
+ "You" (or "Your") shall mean an individual or Legal Entity
+ exercising permissions granted by this License.
+
+ "Source" form shall mean the preferred form for making modifications,
+ including but not limited to software source code, documentation
+ source, and configuration files.
+
+ "Object" form shall mean any form resulting from mechanical
+ transformation or translation of a Source form, including but
+ not limited to compiled object code, generated documentation,
+ and conversions to other media types.
+
+ "Work" shall mean the work of authorship, whether in Source or
+ Object form, made available under the License, as indicated by a
+ copyright notice that is included in or attached to the work
+ (an example is provided in the Appendix below).
+
+ "Derivative Works" shall mean any work, whether in Source or Object
+ form, that is based on (or derived from) the Work and for which the
+ editorial revisions, annotations, elaborations, or other modifications
+ represent, as a whole, an original work of authorship. For the purposes
+ of this License, Derivative Works shall not include works that remain
+ separable from, or merely link (or bind by name) to the interfaces of,
+ the Work and Derivative Works thereof.
+
+ "Contribution" shall mean any work of authorship, including
+ the original version of the Work and any modifications or additions
+ to that Work or Derivative Works thereof, that is intentionally
+ submitted to Licensor for inclusion in the Work by the copyright owner
+ or by an individual or Legal Entity authorized to submit on behalf of
+ the copyright owner. For the purposes of this definition, "submitted"
+ means any form of electronic, verbal, or written communication sent
+ to the Licensor or its representatives, including but not limited to
+ communication on electronic mailing lists, source code control systems,
+ and issue tracking systems that are managed by, or on behalf of, the
+ Licensor for the purpose of discussing and improving the Work, but
+ excluding communication that is conspicuously marked or otherwise
+ designated in writing by the copyright owner as "Not a Contribution."
+
+ "Contributor" shall mean Licensor and any individual or Legal Entity
+ on behalf of whom a Contribution has been received by Licensor and
+ subsequently incorporated within the Work.
+
+ 2. Grant of Copyright License. Subject to the terms and conditions of
+ this License, each Contributor hereby grants to You a perpetual,
+ worldwide, non-exclusive, no-charge, royalty-free, irrevocable
+ copyright license to reproduce, prepare Derivative Works of,
+ publicly display, publicly perform, sublicense, and distribute the
+ Work and such Derivative Works in Source or Object form.
+
+ 3. Grant of Patent License. Subject to the terms and conditions of
+ this License, each Contributor hereby grants to You a perpetual,
+ worldwide, non-exclusive, no-charge, royalty-free, irrevocable
+ (except as stated in this section) patent license to make, have made,
+ use, offer to sell, sell, import, and otherwise transfer the Work,
+ where such license applies only to those patent claims licensable
+ by such Contributor that are necessarily infringed by their
+ Contribution(s) alone or by combination of their Contribution(s)
+ with the Work to which such Contribution(s) was submitted. If You
+ institute patent litigation against any entity (including a
+ cross-claim or counterclaim in a lawsuit) alleging that the Work
+ or a Contribution incorporated within the Work constitutes direct
+ or contributory patent infringement, then any patent licenses
+ granted to You under this License for that Work shall terminate
+ as of the date such litigation is filed.
+
+ 4. Redistribution. You may reproduce and distribute copies of the
+ Work or Derivative Works thereof in any medium, with or without
+ modifications, and in Source or Object form, provided that You
+ meet the following conditions:
+
+ (a) You must give any other recipients of the Work or
+ Derivative Works a copy of this License; and
+
+ (b) You must cause any modified files to carry prominent notices
+ stating that You changed the files; and
+
+ (c) You must retain, in the Source form of any Derivative Works
+ that You distribute, all copyright, patent, trademark, and
+ attribution notices from the Source form of the Work,
+ excluding those notices that do not pertain to any part of
+ the Derivative Works; and
+
+ (d) If the Work includes a "NOTICE" text file as part of its
+ distribution, then any Derivative Works that You distribute must
+ include a readable copy of the attribution notices contained
+ within such NOTICE file, excluding those notices that do not
+ pertain to any part of the Derivative Works, in at least one
+ of the following places: within a NOTICE text file distributed
+ as part of the Derivative Works; within the Source form or
+ documentation, if provided along with the Derivative Works; or,
+ within a display generated by the Derivative Works, if and
+ wherever such third-party notices normally appear. The contents
+ of the NOTICE file are for informational purposes only and
+ do not modify the License. You may add Your own attribution
+ notices within Derivative Works that You distribute, alongside
+ or as an addendum to the NOTICE text from the Work, provided
+ that such additional attribution notices cannot be construed
+ as modifying the License.
+
+ You may add Your own copyright statement to Your modifications and
+ may provide additional or different license terms and conditions
+ for use, reproduction, or distribution of Your modifications, or
+ for any such Derivative Works as a whole, provided Your use,
+ reproduction, and distribution of the Work otherwise complies with
+ the conditions stated in this License.
+
+ 5. Submission of Contributions. Unless You explicitly state otherwise,
+ any Contribution intentionally submitted for inclusion in the Work
+ by You to the Licensor shall be under the terms and conditions of
+ this License, without any additional terms or conditions.
+ Notwithstanding the above, nothing herein shall supersede or modify
+ the terms of any separate license agreement you may have executed
+ with Licensor regarding such Contributions.
+
+ 6. Trademarks. This License does not grant permission to use the trade
+ names, trademarks, service marks, or product names of the Licensor,
+ except as required for reasonable and customary use in describing the
+ origin of the Work and reproducing the content of the NOTICE file.
+
+ 7. Disclaimer of Warranty. Unless required by applicable law or
+ agreed to in writing, Licensor provides the Work (and each
+ Contributor provides its Contributions) on an "AS IS" BASIS,
+ WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
+ implied, including, without limitation, any warranties or conditions
+ of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
+ PARTICULAR PURPOSE. You are solely responsible for determining the
+ appropriateness of using or redistributing the Work and assume any
+ risks associated with Your exercise of permissions under this License.
+
+ 8. Limitation of Liability. In no event and under no legal theory,
+ whether in tort (including negligence), contract, or otherwise,
+ unless required by applicable law (such as deliberate and grossly
+ negligent acts) or agreed to in writing, shall any Contributor be
+ liable to You for damages, including any direct, indirect, special,
+ incidental, or consequential damages of any character arising as a
+ result of this License or out of the use or inability to use the
+ Work (including but not limited to damages for loss of goodwill,
+ work stoppage, computer failure or malfunction, or any and all
+ other commercial damages or losses), even if such Contributor
+ has been advised of the possibility of such damages.
+
+ 9. Accepting Warranty or Additional Liability. While redistributing
+ the Work or Derivative Works thereof, You may choose to offer,
+ and charge a fee for, acceptance of support, warranty, indemnity,
+ or other liability obligations and/or rights consistent with this
+ License. However, in accepting such obligations, You may act only
+ on Your own behalf and on Your sole responsibility, not on behalf
+ of any other Contributor, and only if You agree to indemnify,
+ defend, and hold each Contributor harmless for any liability
+ incurred by, or claims asserted against, such Contributor by reason
+ of your accepting any such warranty or additional liability.
+
+ END OF TERMS AND CONDITIONS
+
+ APPENDIX: How to apply the Apache License to your work.
+
+ To apply the Apache License to your work, attach the following
+ boilerplate notice, with the fields enclosed by brackets "[]"
+ replaced with your own identifying information. (Don't include
+ the brackets!) The text should be enclosed in the appropriate
+ comment syntax for the file format. We also recommend that a
+ file or class name and description of purpose be included on the
+ same "printed page" as the copyright notice for easier
+ identification within third-party archives.
+
+ Copyright [yyyy] [name of copyright owner]
+
+ Licensed under the Apache License, Version 2.0 (the "License");
+ you may not use this file except in compliance with the License.
+ You may obtain a copy of the License at
+
+ http://www.apache.org/licenses/LICENSE-2.0
+
+ Unless required by applicable law or agreed to in writing, software
+ distributed under the License is distributed on an "AS IS" BASIS,
+ WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ See the License for the specific language governing permissions and
+ limitations under the License.
+
+------------------------------------------------------------------------------
+
+Model Weights License
+
+This license applies to model weights in the directory.
+
+Attribution-NonCommercial-ShareAlike 4.0 International
+
+=======================================================================
+
+Creative Commons Corporation ("Creative Commons") is not a law firm and
+does not provide legal services or legal advice. Distribution of
+Creative Commons public licenses does not create a lawyer-client or
+other relationship. Creative Commons makes its licenses and related
+information available on an "as-is" basis. Creative Commons gives no
+warranties regarding its licenses, any material licensed under their
+terms and conditions, or any related information. Creative Commons
+disclaims all liability for damages resulting from their use to the
+fullest extent possible.
+
+Using Creative Commons Public Licenses
+
+Creative Commons public licenses provide a standard set of terms and
+conditions that creators and other rights holders may use to share
+original works of authorship and other material subject to copyright
+and certain other rights specified in the public license below. The
+following considerations are for informational purposes only, are not
+exhaustive, and do not form part of our licenses.
+
+ Considerations for licensors: Our public licenses are
+ intended for use by those authorized to give the public
+ permission to use material in ways otherwise restricted by
+ copyright and certain other rights. Our licenses are
+ irrevocable. Licensors should read and understand the terms
+ and conditions of the license they choose before applying it.
+ Licensors should also secure all rights necessary before
+ applying our licenses so that the public can reuse the
+ material as expected. Licensors should clearly mark any
+ material not subject to the license. This includes other CC-
+ licensed material, or material used under an exception or
+ limitation to copyright. More considerations for licensors:
+ wiki.creativecommons.org/Considerations_for_licensors
+
+ Considerations for the public: By using one of our public
+ licenses, a licensor grants the public permission to use the
+ licensed material under specified terms and conditions. If
+ the licensor's permission is not necessary for any reason--for
+ example, because of any applicable exception or limitation to
+ copyright--then that use is not regulated by the license. Our
+ licenses grant only permissions under copyright and certain
+ other rights that a licensor has authority to grant. Use of
+ the licensed material may still be restricted for other
+ reasons, including because others have copyright or other
+ rights in the material. A licensor may make special requests,
+ such as asking that all changes be marked or described.
+ Although not required by our licenses, you are encouraged to
+ respect those requests where reasonable. More considerations
+ for the public:
+ wiki.creativecommons.org/Considerations_for_licensees
+
+=======================================================================
+
+Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International
+Public License
+
+By exercising the Licensed Rights (defined below), You accept and agree
+to be bound by the terms and conditions of this Creative Commons
+Attribution-NonCommercial-ShareAlike 4.0 International Public License
+("Public License"). To the extent this Public License may be
+interpreted as a contract, You are granted the Licensed Rights in
+consideration of Your acceptance of these terms and conditions, and the
+Licensor grants You such rights in consideration of benefits the
+Licensor receives from making the Licensed Material available under
+these terms and conditions.
+
+
+Section 1 -- Definitions.
+
+ a. Adapted Material means material subject to Copyright and Similar
+ Rights that is derived from or based upon the Licensed Material
+ and in which the Licensed Material is translated, altered,
+ arranged, transformed, or otherwise modified in a manner requiring
+ permission under the Copyright and Similar Rights held by the
+ Licensor. For purposes of this Public License, where the Licensed
+ Material is a musical work, performance, or sound recording,
+ Adapted Material is always produced where the Licensed Material is
+ synched in timed relation with a moving image.
+
+ b. Adapter's License means the license You apply to Your Copyright
+ and Similar Rights in Your contributions to Adapted Material in
+ accordance with the terms and conditions of this Public License.
+
+ c. BY-NC-SA Compatible License means a license listed at
+ creativecommons.org/compatiblelicenses, approved by Creative
+ Commons as essentially the equivalent of this Public License.
+
+ d. Copyright and Similar Rights means copyright and/or similar rights
+ closely related to copyright including, without limitation,
+ performance, broadcast, sound recording, and Sui Generis Database
+ Rights, without regard to how the rights are labeled or
+ categorized. For purposes of this Public License, the rights
+ specified in Section 2(b)(1)-(2) are not Copyright and Similar
+ Rights.
+
+ e. Effective Technological Measures means those measures that, in the
+ absence of proper authority, may not be circumvented under laws
+ fulfilling obligations under Article 11 of the WIPO Copyright
+ Treaty adopted on December 20, 1996, and/or similar international
+ agreements.
+
+ f. Exceptions and Limitations means fair use, fair dealing, and/or
+ any other exception or limitation to Copyright and Similar Rights
+ that applies to Your use of the Licensed Material.
+
+ g. License Elements means the license attributes listed in the name
+ of a Creative Commons Public License. The License Elements of this
+ Public License are Attribution, NonCommercial, and ShareAlike.
+
+ h. Licensed Material means the artistic or literary work, database,
+ or other material to which the Licensor applied this Public
+ License.
+
+ i. Licensed Rights means the rights granted to You subject to the
+ terms and conditions of this Public License, which are limited to
+ all Copyright and Similar Rights that apply to Your use of the
+ Licensed Material and that the Licensor has authority to license.
+
+ j. Licensor means the individual(s) or entity(ies) granting rights
+ under this Public License.
+
+ k. NonCommercial means not primarily intended for or directed towards
+ commercial advantage or monetary compensation. For purposes of
+ this Public License, the exchange of the Licensed Material for
+ other material subject to Copyright and Similar Rights by digital
+ file-sharing or similar means is NonCommercial provided there is
+ no payment of monetary compensation in connection with the
+ exchange.
+
+ l. Share means to provide material to the public by any means or
+ process that requires permission under the Licensed Rights, such
+ as reproduction, public display, public performance, distribution,
+ dissemination, communication, or importation, and to make material
+ available to the public including in ways that members of the
+ public may access the material from a place and at a time
+ individually chosen by them.
+
+ m. Sui Generis Database Rights means rights other than copyright
+ resulting from Directive 96/9/EC of the European Parliament and of
+ the Council of 11 March 1996 on the legal protection of databases,
+ as amended and/or succeeded, as well as other essentially
+ equivalent rights anywhere in the world.
+
+ n. You means the individual or entity exercising the Licensed Rights
+ under this Public License. Your has a corresponding meaning.
+
+
+Section 2 -- Scope.
+
+ a. License grant.
+
+ 1. Subject to the terms and conditions of this Public License,
+ the Licensor hereby grants You a worldwide, royalty-free,
+ non-sublicensable, non-exclusive, irrevocable license to
+ exercise the Licensed Rights in the Licensed Material to:
+
+ a. reproduce and Share the Licensed Material, in whole or
+ in part, for NonCommercial purposes only; and
+
+ b. produce, reproduce, and Share Adapted Material for
+ NonCommercial purposes only.
+
+ 2. Exceptions and Limitations. For the avoidance of doubt, where
+ Exceptions and Limitations apply to Your use, this Public
+ License does not apply, and You do not need to comply with
+ its terms and conditions.
+
+ 3. Term. The term of this Public License is specified in Section
+ 6(a).
+
+ 4. Media and formats; technical modifications allowed. The
+ Licensor authorizes You to exercise the Licensed Rights in
+ all media and formats whether now known or hereafter created,
+ and to make technical modifications necessary to do so. The
+ Licensor waives and/or agrees not to assert any right or
+ authority to forbid You from making technical modifications
+ necessary to exercise the Licensed Rights, including
+ technical modifications necessary to circumvent Effective
+ Technological Measures. For purposes of this Public License,
+ simply making modifications authorized by this Section 2(a)
+ (4) never produces Adapted Material.
+
+ 5. Downstream recipients.
+
+ a. Offer from the Licensor -- Licensed Material. Every
+ recipient of the Licensed Material automatically
+ receives an offer from the Licensor to exercise the
+ Licensed Rights under the terms and conditions of this
+ Public License.
+
+ b. Additional offer from the Licensor -- Adapted Material.
+ Every recipient of Adapted Material from You
+ automatically receives an offer from the Licensor to
+ exercise the Licensed Rights in the Adapted Material
+ under the conditions of the Adapter's License You apply.
+
+ c. No downstream restrictions. You may not offer or impose
+ any additional or different terms or conditions on, or
+ apply any Effective Technological Measures to, the
+ Licensed Material if doing so restricts exercise of the
+ Licensed Rights by any recipient of the Licensed
+ Material.
+
+ 6. No endorsement. Nothing in this Public License constitutes or
+ may be construed as permission to assert or imply that You
+ are, or that Your use of the Licensed Material is, connected
+ with, or sponsored, endorsed, or granted official status by,
+ the Licensor or others designated to receive attribution as
+ provided in Section 3(a)(1)(A)(i).
+
+ b. Other rights.
+
+ 1. Moral rights, such as the right of integrity, are not
+ licensed under this Public License, nor are publicity,
+ privacy, and/or other similar personality rights; however, to
+ the extent possible, the Licensor waives and/or agrees not to
+ assert any such rights held by the Licensor to the limited
+ extent necessary to allow You to exercise the Licensed
+ Rights, but not otherwise.
+
+ 2. Patent and trademark rights are not licensed under this
+ Public License.
+
+ 3. To the extent possible, the Licensor waives any right to
+ collect royalties from You for the exercise of the Licensed
+ Rights, whether directly or through a collecting society
+ under any voluntary or waivable statutory or compulsory
+ licensing scheme. In all other cases the Licensor expressly
+ reserves any right to collect such royalties, including when
+ the Licensed Material is used other than for NonCommercial
+ purposes.
+
+
+Section 3 -- License Conditions.
+
+Your exercise of the Licensed Rights is expressly made subject to the
+following conditions.
+
+ a. Attribution.
+
+ 1. If You Share the Licensed Material (including in modified
+ form), You must:
+
+ a. retain the following if it is supplied by the Licensor
+ with the Licensed Material:
+
+ i. identification of the creator(s) of the Licensed
+ Material and any others designated to receive
+ attribution, in any reasonable manner requested by
+ the Licensor (including by pseudonym if
+ designated);
+
+ ii. a copyright notice;
+
+ iii. a notice that refers to this Public License;
+
+ iv. a notice that refers to the disclaimer of
+ warranties;
+
+ v. a URI or hyperlink to the Licensed Material to the
+ extent reasonably practicable;
+
+ b. indicate if You modified the Licensed Material and
+ retain an indication of any previous modifications; and
+
+ c. indicate the Licensed Material is licensed under this
+ Public License, and include the text of, or the URI or
+ hyperlink to, this Public License.
+
+ 2. You may satisfy the conditions in Section 3(a)(1) in any
+ reasonable manner based on the medium, means, and context in
+ which You Share the Licensed Material. For example, it may be
+ reasonable to satisfy the conditions by providing a URI or
+ hyperlink to a resource that includes the required
+ information.
+ 3. If requested by the Licensor, You must remove any of the
+ information required by Section 3(a)(1)(A) to the extent
+ reasonably practicable.
+
+ b. ShareAlike.
+
+ In addition to the conditions in Section 3(a), if You Share
+ Adapted Material You produce, the following conditions also apply.
+
+ 1. The Adapter's License You apply must be a Creative Commons
+ license with the same License Elements, this version or
+ later, or a BY-NC-SA Compatible License.
+
+ 2. You must include the text of, or the URI or hyperlink to, the
+ Adapter's License You apply. You may satisfy this condition
+ in any reasonable manner based on the medium, means, and
+ context in which You Share Adapted Material.
+
+ 3. You may not offer or impose any additional or different terms
+ or conditions on, or apply any Effective Technological
+ Measures to, Adapted Material that restrict exercise of the
+ rights granted under the Adapter's License You apply.
+
+
+Section 4 -- Sui Generis Database Rights.
+
+Where the Licensed Rights include Sui Generis Database Rights that
+apply to Your use of the Licensed Material:
+
+ a. for the avoidance of doubt, Section 2(a)(1) grants You the right
+ to extract, reuse, reproduce, and Share all or a substantial
+ portion of the contents of the database for NonCommercial purposes
+ only;
+
+ b. if You include all or a substantial portion of the database
+ contents in a database in which You have Sui Generis Database
+ Rights, then the database in which You have Sui Generis Database
+ Rights (but not its individual contents) is Adapted Material,
+ including for purposes of Section 3(b); and
+
+ c. You must comply with the conditions in Section 3(a) if You Share
+ all or a substantial portion of the contents of the database.
+
+For the avoidance of doubt, this Section 4 supplements and does not
+replace Your obligations under this Public License where the Licensed
+Rights include other Copyright and Similar Rights.
+
+
+Section 5 -- Disclaimer of Warranties and Limitation of Liability.
+
+ a. UNLESS OTHERWISE SEPARATELY UNDERTAKEN BY THE LICENSOR, TO THE
+ EXTENT POSSIBLE, THE LICENSOR OFFERS THE LICENSED MATERIAL AS-IS
+ AND AS-AVAILABLE, AND MAKES NO REPRESENTATIONS OR WARRANTIES OF
+ ANY KIND CONCERNING THE LICENSED MATERIAL, WHETHER EXPRESS,
+ IMPLIED, STATUTORY, OR OTHER. THIS INCLUDES, WITHOUT LIMITATION,
+ WARRANTIES OF TITLE, MERCHANTABILITY, FITNESS FOR A PARTICULAR
+ PURPOSE, NON-INFRINGEMENT, ABSENCE OF LATENT OR OTHER DEFECTS,
+ ACCURACY, OR THE PRESENCE OR ABSENCE OF ERRORS, WHETHER OR NOT
+ KNOWN OR DISCOVERABLE. WHERE DISCLAIMERS OF WARRANTIES ARE NOT
+ ALLOWED IN FULL OR IN PART, THIS DISCLAIMER MAY NOT APPLY TO YOU.
+
+ b. TO THE EXTENT POSSIBLE, IN NO EVENT WILL THE LICENSOR BE LIABLE
+ TO YOU ON ANY LEGAL THEORY (INCLUDING, WITHOUT LIMITATION,
+ NEGLIGENCE) OR OTHERWISE FOR ANY DIRECT, SPECIAL, INDIRECT,
+ INCIDENTAL, CONSEQUENTIAL, PUNITIVE, EXEMPLARY, OR OTHER LOSSES,
+ COSTS, EXPENSES, OR DAMAGES ARISING OUT OF THIS PUBLIC LICENSE OR
+ USE OF THE LICENSED MATERIAL, EVEN IF THE LICENSOR HAS BEEN
+ ADVISED OF THE POSSIBILITY OF SUCH LOSSES, COSTS, EXPENSES, OR
+ DAMAGES. WHERE A LIMITATION OF LIABILITY IS NOT ALLOWED IN FULL OR
+ IN PART, THIS LIMITATION MAY NOT APPLY TO YOU.
+
+ c. The disclaimer of warranties and limitation of liability provided
+ above shall be interpreted in a manner that, to the extent
+ possible, most closely approximates an absolute disclaimer and
+ waiver of all liability.
+
+
+Section 6 -- Term and Termination.
+
+ a. This Public License applies for the term of the Copyright and
+ Similar Rights licensed here. However, if You fail to comply with
+ this Public License, then Your rights under this Public License
+ terminate automatically.
+
+ b. Where Your right to use the Licensed Material has terminated under
+ Section 6(a), it reinstates:
+
+ 1. automatically as of the date the violation is cured, provided
+ it is cured within 30 days of Your discovery of the
+ violation; or
+
+ 2. upon express reinstatement by the Licensor.
+
+ For the avoidance of doubt, this Section 6(b) does not affect any
+ right the Licensor may have to seek remedies for Your violations
+ of this Public License.
+
+ c. For the avoidance of doubt, the Licensor may also offer the
+ Licensed Material under separate terms or conditions or stop
+ distributing the Licensed Material at any time; however, doing so
+ will not terminate this Public License.
+
+ d. Sections 1, 5, 6, 7, and 8 survive termination of this Public
+ License.
+
+
+Section 7 -- Other Terms and Conditions.
+
+ a. The Licensor shall not be bound by any additional or different
+ terms or conditions communicated by You unless expressly agreed.
+
+ b. Any arrangements, understandings, or agreements regarding the
+ Licensed Material not stated herein are separate from and
+ independent of the terms and conditions of this Public License.
+
+
+Section 8 -- Interpretation.
+
+ a. For the avoidance of doubt, this Public License does not, and
+ shall not be interpreted to, reduce, limit, restrict, or impose
+ conditions on any use of the Licensed Material that could lawfully
+ be made without permission under this Public License.
+
+ b. To the extent possible, if any provision of this Public License is
+ deemed unenforceable, it shall be automatically reformed to the
+ minimum extent necessary to make it enforceable. If the provision
+ cannot be reformed, it shall be severed from this Public License
+ without affecting the enforceability of the remaining terms and
+ conditions.
+
+ c. No term or condition of this Public License will be waived and no
+ failure to comply consented to unless expressly agreed to by the
+ Licensor.
+
+ d. Nothing in this Public License constitutes or may be interpreted
+ as a limitation upon, or waiver of, any privileges and immunities
+ that apply to the Licensor or You, including from the legal
+ processes of any jurisdiction or authority.
+
+=======================================================================
+
+Creative Commons is not a party to its public
+licenses. Notwithstanding, Creative Commons may elect to apply one of
+its public licenses to material it publishes and in those instances
+will be considered the “Licensor.” The text of the Creative Commons
+public licenses is dedicated to the public domain under the CC0 Public
+Domain Dedication. Except for the limited purpose of indicating that
+material is shared under a Creative Commons public license or as
+otherwise permitted by the Creative Commons policies published at
+creativecommons.org/policies, Creative Commons does not authorize the
+use of the trademark "Creative Commons" or any other trademark or logo
+of Creative Commons without its prior written consent including,
+without limitation, in connection with any unauthorized modifications
+to any of its public licenses or any other arrangements,
+understandings, or agreements concerning use of licensed material. For
+the avoidance of doubt, this paragraph does not form part of the
+public licenses.
+
+Creative Commons may be contacted at creativecommons.org.
diff --git a/models/vista2d/configs/hyper_parameters.yaml b/models/vista2d/configs/hyper_parameters.yaml
new file mode 100644
index 00000000..b05fce30
--- /dev/null
+++ b/models/vista2d/configs/hyper_parameters.yaml
@@ -0,0 +1,135 @@
+imports:
+ - $import os
+
+# seed: 28022024 # uncommend for deterministic results (but slower)
+seed: null
+
+bundle_root: "."
+ckpt_path: $os.path.join(@bundle_root, "models") # location to save checkpoints
+output_dir: $os.path.join(@bundle_root, "eval") # location to save events and logs
+log_output_file: $os.path.join(@output_dir, "vista_cell.log")
+
+mlflow_tracking_uri: null # enable mlflow logging, e.g. $@ckpt_path + '/mlruns/ or "http://127.0.0.1:8080" or a remote url
+mlflow_log_system_metrics: true # log system metrics to mlflow (requires: pip install psutil pynvml)
+mlflow_run_name: null # optional name of the current run
+
+ckpt_save: true # save checkpoints periodically
+amp: true
+amp_dtype: "float16" #float16 or bfloat16 (Ampere or newer)
+channels_last: true
+compile: false # complie the model for faster processing
+
+start_epoch: 0
+run_final_testing: true
+use_weighted_sampler: false # only applicable when using several dataset jsons for data_list_files
+
+pretrained_ckpt_name: null
+pretrained_ckpt_path: null
+
+# for commandline setting of a single dataset
+datalist: datalists/cellpose_datalist.json
+basedir: /cellpose_dataset
+data_list_files:
+ - {datalist: "@datalist", basedir: "@basedir"}
+
+
+fold: 0
+learning_rate: 0.01 # try 1.0e-4 if using AdamW
+quick: false # whether to use a small subset of data for quick testing
+roi_size: [256, 256]
+
+train:
+ skip: false
+ handlers: []
+ trainer:
+ num_warmup_epochs: 3
+ max_epochs: 200
+ num_epochs_per_saving: 1
+ num_epochs_per_validation: null
+ num_workers: 4
+ batch_size: 1
+ dataset:
+ preprocessing:
+ roi_size: "@roi_size"
+ data:
+ key: null # set to 'testing' to use this subset in periodic validations, instead of the the validation set
+ data_list_files: "@data_list_files"
+
+dataset:
+ data:
+ key: "testing"
+ data_list_files: "@data_list_files"
+
+validate:
+ grouping: true
+ evaluator:
+ postprocessing: "@postprocessing"
+ dataset:
+ data: "@dataset#data"
+ batch_size: 1
+ num_workers: 4
+ preprocessing: null
+ postprocessing: null
+ inferer: null
+ handlers: null
+ key_metric: null
+
+infer:
+ evaluator:
+ postprocessing: "@postprocessing"
+ dataset:
+ data: "@dataset#data"
+
+
+device: "$torch.device(('cuda:' + os.environ.get('LOCAL_RANK', '0')) if torch.cuda.is_available() else 'cpu')"
+network_def:
+ _target_: scripts.cell_sam_wrapper.CellSamWrapper
+ checkpoint: $os.path.join(@ckpt_path, "sam_vit_b_01ec64.pth")
+network: $@network_def.to(@device)
+
+loss_function:
+ _target_: scripts.components.CellLoss
+
+key_metric:
+ _target_: scripts.components.CellAcc
+
+# optimizer:
+# _target_: torch.optim.AdamW
+# params: $@network.parameters()
+# lr: "@learning_rate"
+# weight_decay: 1.0e-5
+
+optimizer:
+ _target_: torch.optim.SGD
+ params: $@network.parameters()
+ momentum: 0.9
+ lr: "@learning_rate"
+ weight_decay: 1.0e-5
+
+lr_scheduler:
+ _target_: monai.optimizers.lr_scheduler.WarmupCosineSchedule
+ optimizer: "@optimizer"
+ warmup_steps: "@train#trainer#num_warmup_epochs"
+ warmup_multiplier: 0.1
+ t_total: "@train#trainer#max_epochs"
+
+inferer:
+ sliding_inferer:
+ _target_: monai.inferers.SlidingWindowInfererAdapt
+ roi_size: "@roi_size"
+ sw_batch_size: 1
+ overlap: 0.625
+ mode: "gaussian"
+ cache_roi_weight_map: true
+ progress: false
+
+image_saver:
+ _target_: scripts.components.SaveTiffd
+ keys: "seg"
+ output_dir: "@output_dir"
+ nested_folder: false
+
+postprocessing:
+ _target_: monai.transforms.Compose
+ transforms:
+ - "@image_saver"
diff --git a/models/vista2d/configs/inference.json b/models/vista2d/configs/inference.json
new file mode 100644
index 00000000..87dd4f3e
--- /dev/null
+++ b/models/vista2d/configs/inference.json
@@ -0,0 +1,133 @@
+{
+ "imports": [
+ "$import numpy as np"
+ ],
+ "bundle_root": ".",
+ "ckpt_dir": "$@bundle_root + '/models'",
+ "output_dir": "$@bundle_root + '/eval'",
+ "output_ext": ".tif",
+ "output_postfix": "trans",
+ "roi_size": [
+ 256,
+ 256
+ ],
+ "input_dict": "${'image': '/home/venn/Desktop/data/medical/cellpose_dataset/test/001_img.png'}",
+ "device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
+ "sam_ckpt_path": "$@ckpt_dir + '/sam_vit_b_01ec64.pth'",
+ "pretrained_ckpt_path": "$@ckpt_dir + '/model.pt'",
+ "image_key": "image",
+ "channels_last": true,
+ "use_amp": true,
+ "amp_dtype": "$torch.float",
+ "network_def": {
+ "_target_": "scripts.cell_sam_wrapper.CellSamWrapper",
+ "checkpoint": "@sam_ckpt_path"
+ },
+ "network": "$@network_def.to(@device)",
+ "preprocessing_transforms": [
+ {
+ "_target_": "scripts.components.LoadTiffd",
+ "keys": "@image_key"
+ },
+ {
+ "_target_": "EnsureTyped",
+ "keys": "@image_key",
+ "data_type": "tensor",
+ "dtype": "$torch.float"
+ },
+ {
+ "_target_": "ScaleIntensityd",
+ "keys": "@image_key",
+ "minv": 0,
+ "maxv": 1,
+ "channel_wise": true
+ },
+ {
+ "_target_": "ScaleIntensityRangePercentilesd",
+ "keys": "image",
+ "lower": 1,
+ "upper": 99,
+ "b_min": 0.0,
+ "b_max": 1.0,
+ "channel_wise": true,
+ "clip": true
+ }
+ ],
+ "preprocessing": {
+ "_target_": "Compose",
+ "transforms": "$@preprocessing_transforms "
+ },
+ "dataset": {
+ "_target_": "Dataset",
+ "data": "$[@input_dict]",
+ "transform": "@preprocessing"
+ },
+ "dataloader": {
+ "_target_": "ThreadDataLoader",
+ "dataset": "@dataset",
+ "batch_size": 1,
+ "shuffle": false,
+ "num_workers": 0
+ },
+ "inferer": {
+ "_target_": "SlidingWindowInfererAdapt",
+ "roi_size": "@roi_size",
+ "sw_batch_size": 1,
+ "overlap": 0.625,
+ "mode": "gaussian",
+ "cache_roi_weight_map": true,
+ "progress": false
+ },
+ "postprocessing": {
+ "_target_": "Compose",
+ "transforms": [
+ {
+ "_target_": "ToDeviced",
+ "keys": "pred",
+ "device": "cpu"
+ },
+ {
+ "_target_": "scripts.components.LogitsToLabelsd",
+ "keys": "pred"
+ },
+ {
+ "_target_": "scripts.components.SaveTiffExd",
+ "keys": "pred",
+ "output_dir": "@output_dir",
+ "output_ext": "@output_ext",
+ "output_postfix": "@output_postfix"
+ }
+ ]
+ },
+ "handlers": [
+ {
+ "_target_": "StatsHandler",
+ "iteration_log": false
+ }
+ ],
+ "checkpointloader": {
+ "_target_": "CheckpointLoader",
+ "load_path": "@pretrained_ckpt_path",
+ "map_location": "cpu",
+ "load_dict": {
+ "state_dict": "@network"
+ }
+ },
+ "evaluator": {
+ "_target_": "SupervisedEvaluator",
+ "device": "@device",
+ "val_data_loader": "@dataloader",
+ "network": "@network",
+ "inferer": "@inferer",
+ "postprocessing": "@postprocessing",
+ "val_handlers": "@handlers",
+ "amp": true
+ },
+ "initialize": [
+ "$monai.utils.set_determinism(seed=123)",
+ "$@checkpointloader(@evaluator)"
+ ],
+ "run": [
+ "$@evaluator.run()"
+ ]
+}
diff --git a/models/vista2d/configs/metadata.json b/models/vista2d/configs/metadata.json
new file mode 100644
index 00000000..f256392f
--- /dev/null
+++ b/models/vista2d/configs/metadata.json
@@ -0,0 +1,87 @@
+{
+ "schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20240725.json",
+ "version": "0.2.1",
+ "changelog": {
+ "0.2.1": "initial OSS version"
+ },
+ "monai_version": "1.4.0",
+ "pytorch_version": "2.4.0",
+ "numpy_version": "1.24.4",
+ "required_packages_version": {
+ "einops": "0.7.0",
+ "scikit-image": "0.23.2",
+ "cucim-cu12": "24.6.0",
+ "gdown": "5.2.0",
+ "fire": "0.6.0",
+ "pyyaml": "6.0.1",
+ "tensorboard": "2.17.0",
+ "opencv-python": "4.7.0.68",
+ "numba": "0.59.1",
+ "torchvision": "0.19.0",
+ "cellpose": "3.0.8",
+ "natsort": "8.4.0",
+ "roifile": "2024.5.24",
+ "tifffile": "2024.7.2",
+ "fastremap": "1.15.0",
+ "imagecodecs": "2024.6.1",
+ "segment_anything": "1.0"
+ },
+ "optional_packages_version": {
+ "mlflow": "2.14.3",
+ "pynvml": "11.4.1",
+ "psutil": "5.9.8"
+ },
+ "supported_apps": {},
+ "name": "VISTA-Cell",
+ "task": "cell image segmentation",
+ "description": "VISTA2D bundle for cell image analysis",
+ "authors": "MONAI team",
+ "copyright": "Copyright (c) MONAI Consortium",
+ "data_type": "tiff",
+ "image_classes": "1 channel data, intensity scaled to [0, 1]",
+ "label_classes": "3-channel data",
+ "pred_classes": "3 channels",
+ "eval_metrics": {
+ "mean_dice": 0.0
+ },
+ "intended_use": "This is an example, not to be used for diagnostic purposes",
+ "references": [],
+ "network_data_format": {
+ "inputs": {
+ "image": {
+ "type": "image",
+ "num_channels": 3,
+ "spatial_shape": [
+ 256,
+ 256
+ ],
+ "format": "RGB",
+ "value_range": [
+ 0,
+ 255
+ ],
+ "dtype": "float32",
+ "is_patch_data": true,
+ "channel_def": {
+ "0": "image"
+ }
+ }
+ },
+ "outputs": {
+ "pred": {
+ "type": "image",
+ "format": "segmentation",
+ "num_channels": 3,
+ "dtype": "float32",
+ "value_range": [
+ 0,
+ 1
+ ],
+ "spatial_shape": [
+ 256,
+ 256
+ ]
+ }
+ }
+ }
+}
diff --git a/models/vista2d/docs/README.md b/models/vista2d/docs/README.md
new file mode 100644
index 00000000..c2f221be
--- /dev/null
+++ b/models/vista2d/docs/README.md
@@ -0,0 +1,169 @@
+## Overview
+
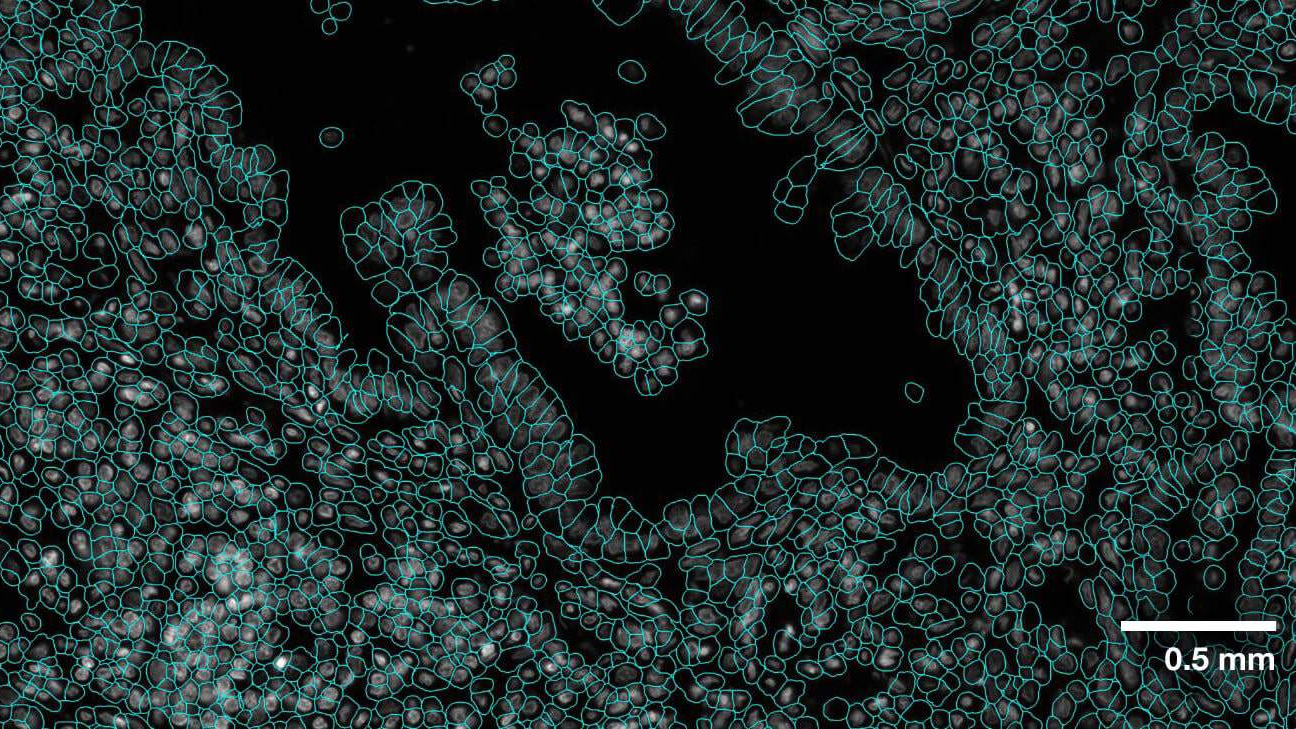
+The **VISTA2D** is a cell segmentation training and inference pipeline for cell imaging [[`Blog`](https://developer.nvidia.com/blog/advancing-cell-segmentation-and-morphology-analysis-with-nvidia-ai-foundation-model-vista-2d/)].
+
+A pretrained model was trained on collection of 15K public microscopy images. The data collection and training can be reproduced following the [tutorial](../download_preprocessor/). Alternatively, the model can be retrained on your own dataset. The pretrained vista2d model achieves good performance on diverse set of cell types, microscopy image modalities, and can be further finetuned if necessary. The codebase utilizes several components from other great works including [SegmentAnything](https://github.com/facebookresearch/segment-anything) and [Cellpose](https://www.cellpose.org/), which must be pip installed as dependencies. Vista2D codebase follows MONAI bundle format and its [specifications](https://docs.monai.io/en/stable/mb_specification.html).
+
+ 
+
+
+### Model highlights
+
+- Robust deep learning algorithm based on transformers
+- Generalist model as compared to specialist models
+- Multiple dataset sources and file formats supported
+- Multiple modalities of imaging data collectively supported
+- Multi-GPU and multinode training support
+
+
+### Generalization performance
+
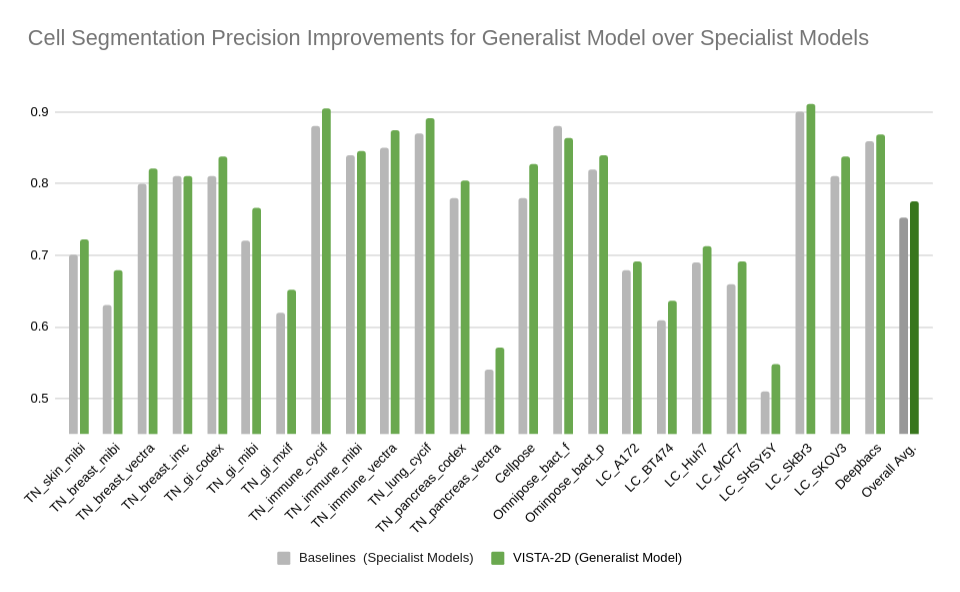
+Evaluation was performed for the VISTA2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and [more](./docs/data_license.txt). A total of ~15K annotated cell images were collected to train the generalist VISTA2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types. The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. Average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA2D model trained only on a particular dataset or a subset of data.
+
+ 
+
+
+### Prepare Data Lists and Datasets
+
+The default dataset for training, validation, and inference is the [Cellpose](https://www.cellpose.org/) dataset. Please follow the [tutorial](../download_preprocessor/) to prepare the dataset before executing any commands below.
+
+Additionally, all data lists are available in the `datalists.zip` file located in the root directory of the bundle. Extract the contents of the `.zip` file to access the data lists.
+
+### Execute training
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml
+```
+
+You can override the `basedir` to specify a different dataset directory by using the following command:
+
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --basedir
+```
+
+#### Quick run with a few data points
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --quick True --train#trainer#max_epochs 3
+```
+
+### Execute multi-GPU training
+```bash
+torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml
+```
+
+### Execute validation
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --pretrained_ckpt_name model.pt --mode eval
+```
+(can append `--quick True` for quick demoing)
+
+### Execute multi-GPU validation
+```bash
+torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode eval
+```
+
+### Execute inference
+```bash
+python -m monai.bundle run --config_file configs/inference.json
+```
+
+Please note that the data used in the config file is: "/cellpose_dataset/test/001_img.png", if the dataset path is different or you want to do inference on another file, please modify in `configs/inference.json` accordingly.
+
+### Execute multi-GPU inference
+```bash
+torchrun --nproc_per_node=gpu -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mode infer --pretrained_ckpt_name model.pt
+```
+(can append `--quick True` for quick demoing)
+
+
+
+#### Finetune starting from a trained checkpoint
+(we use a smaller learning rate, small number of epochs, and initialize from a checkpoint)
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --learning_rate=0.001 --train#trainer#max_epochs 20 --pretrained_ckpt_path /path/to/saved/model.pt
+```
+
+
+#### Configuration options
+
+To disable the segmentation writing:
+```
+--postprocessing []
+```
+
+Load a checkpoint for validation or inference (relative path within results directory):
+```
+--pretrained_ckpt_name "model.pt"
+```
+
+Load a checkpoint for validation or inference (absolute path):
+```
+--pretrained_ckpt_path "/path/to/another/location/model.pt"
+```
+
+`--mode eval` or `--mode infer`will use the corresponding configurations from the `validate` or `infer`
+of the `configs/hyper_parameters.yaml`.
+
+By default the generated `model.pt` corresponds to the checkpoint at the best validation score,
+`model_final.pt` is the checkpoint after the latest training epoch.
+
+
+### Development
+
+For development purposes it's possible to run the script directly (without monai bundle calls)
+
+```bash
+python scripts/workflow.py --config_file configs/hyper_parameters.yaml ...
+torchrun --nproc_per_node=gpu -m scripts/workflow.py --config_file configs/hyper_parameters.yaml ..
+```
+
+### MLFlow support
+
+Enable MLFlow logging by specifying "mlflow_tracking_uri" (can be local or remote URL).
+
+```bash
+python -m monai.bundle run_workflow "scripts.workflow.VistaCell" --config_file configs/hyper_parameters.yaml --mlflow_tracking_uri=http://127.0.0.1:8080
+```
+
+Optionally use "--mlflow_run_name=.." to specify MLFlow experiment name, and "--mlflow_log_system_metrics=True/False" to enable logging of CPU/GPU resources (requires pip install psutil pynvml)
+
+
+
+### Unit tests
+
+Test single GPU training:
+```
+python unit_tests/test_vista2d.py
+```
+
+Test multi-GPU training (may need to uncomment the `"--standalone"` in the `unit_tests/utils.py` file):
+```
+python unit_tests/test_vista2d_mgpu.py
+```
+
+## Compute Requirements
+Min GPU memory requirements 16Gb.
+
+
+## Contributing
+Vista2D codebase follows MONAI bundle format and its [specifications](https://docs.monai.io/en/stable/mb_specification.html).
+Make sure to run pre-commit before committing code changes to git
+```bash
+pip install pre-commit
+python3 -m pre_commit run --all-files
+```
+
+
+## Community
+
+Join the conversation on Twitter [@ProjectMONAI](https://twitter.com/ProjectMONAI) or join
+our [Slack channel](https://projectmonai.slack.com/archives/C031QRE0M1C).
+
+Ask and answer questions on [MONAI VISTA's GitHub discussions tab](https://github.com/Project-MONAI/VISTA/discussions).
+
+## License
+
+The codebase is under Apache 2.0 Licence. The model weight is released under CC-BY-NC-SA-4.0. For various public data licenses please see [data_license.txt](data_license.txt).
+
+## Acknowledgement
+- [segment-anything](https://github.com/facebookresearch/segment-anything)
+- [Cellpose](https://www.cellpose.org/)
diff --git a/models/vista2d/docs/data_license.txt b/models/vista2d/docs/data_license.txt
new file mode 100644
index 00000000..89fce562
--- /dev/null
+++ b/models/vista2d/docs/data_license.txt
@@ -0,0 +1,361 @@
+Third Party Licenses
+-----------------------------------------------------------------------
+
+/*********************************************************************/
+i.Cellpose dataset
+
+ https://www.cellpose.org/dataset
+ The user agrees to the listed conditions of Cellpose dataset by default that are cited below:
+
+ Howard Hughes Medical Institute
+
+ Research Content Terms and Conditions
+
+ Please read these Research Content Terms and Conditions (“Terms and Conditions”) carefully before you download or use
+ any images in any format from the cellpose.org website (“Content”), and do not download or use Content if you do not
+ agree with these Terms and Conditions. The Howard Hughes Medical Institute (“HHMI”, “we”, “us” and “our”) may at any
+ time revise these Terms and Conditions by updating this posting. You are bound by any such revisions and should
+ therefore periodically visit this page to review the then-current Terms and Conditions.
+
+ BY ACCEPTING THESE TERMS AND CONDITIONS, DOWNLOADING THE CONTENT OR USING THE CONTENT, YOU ARE CONFIRMING YOUR AGREEMENT
+ TO BE BOUND BY THESE TERMS AND CONDITIONS INCLUDING THE WARRANTY DISCLAIMERS, LIMITATIONS OF LIABILITY AND TERMINATION
+ PROVISIONS BELOW. IF ANY OF THESE TERMS AND CONDITIONS OR ANY FUTURE CHANGES ARE UNACCEPTABLE TO YOU, DO NOT DOWNLOAD
+ OR USE THE CONTENT AT THE CELLPOSE.ORG WEBSITE. BY DOWNLOADING OR USING CONTENT FROM CELLPOSE.ORG YOU ACCEPT AND AGREE
+ TO THESE TERMS AND CONDITIONS WITHOUT ANY RESERVATIONS, MODIFICATIONS, ADDITIONS, OR DELETIONS. IF YOU DO NOT AGREE TO
+ THESE TERMS AND CONDITIONS, YOU ARE NOT AUTHORIZED TO DOWNLOAD OR USE THE CONTENT. IF YOU REPRESENT A CORPORATION,
+ PARTNERSHIP, OR OTHER NON-INDIVIDUAL ENTITY, THE PERSON ACCEPTING THESE TERMS AND CONDITIONS ON BEHALF OF THAT ENTITY
+ REPRESENTS AND WARRANTS THAT THEY HAVE ALL NECESSARY AUTHORITY TO BIND THAT ENTITY.
+
+ Ownership
+ All Content is protected by copyright, and such copyrights and other proprietary rights may be held by individuals or
+ entities other than, or in addition to, us.
+ Use and Restrictions
+ The Content is made available for limited non-commercial, educational, research and personal use only, and for fair use
+ as defined under United States copyright laws. You may download and use Content only for your own non-commercial,
+ educational, research and personal use only, subject to any additional terms or restrictions which may be applicable to
+ an individual file as part of the Content. Copying or redistribution of the Content in any manner for commercial use,
+ including commercial publication, or for personal gain, or making any other use of the Content beyond that allowed by
+ “fair use,” as such term is understood under the United States Copyright Act and applicable law, is strictly prohibited.
+ HHMI may terminate these Terms and Conditions, and your right to use the Content at any time upon notice to you (which
+ notice may be to your email address of record with HHMI). Upon any termination by HHMI, you agree that you will promptly
+ delete all copies of Content and, upon request by HHMI, certify in writing your deletion of all copies of Content.
+ Indemnity
+ You agree to indemnify, defend, and hold harmless HHMI and our affiliates, and our trustees, officers, members,
+ directors, employees, representatives and agents from and against all claims, losses, expenses, damages, costs and other
+ liability (including without limitation attorneys’ fees), arising or resulting from your use of the Content (including,
+ without limitation, any copies and derivative works of any Content), or any violation or alleged violation by you of
+ these Terms and Conditions, including for any violation of any applicable law, rule, or regulation. We reserve the
+ right to assume, at our sole expense, the exclusive defense and control of any matter subject to indemnification by
+ you, in which event you will fully cooperate with us.
+
+ Disclaimers
+ WE MAKE NO EXPRESS WARRANTIES OR REPRESENTATIONS AS TO THE QUALITY, COMPREHENSIVENESS, AND ACCURACY OF THE CONTENT, AND
+ WE DISCLAIM ANY IMPLIED WARRANTIES OR REPRESENTATIONS, INCLUDING BUT NOT LIMITED TO IMPLIED WARRANTIES OF MERCHANTABILITY,
+ FITNESS FOR A PARTICULAR PURPOSE, OR NON-INFRINGEMENT, TO THE FULL EXTENT PERMISSIBLE UNDER APPLICABLE LAW. WE OFFER THE
+ CONTENT ON AN "AS IS” BASIS AND DO NOT ACCEPT RESPONSIBILITY FOR ANY USE OF OR RELIANCE ON THE CONTENT. IN ADDITION, WE
+ DO NOT MAKE ANY REPRESENTATIONS AS TO THE ACCURACY, COMPREHENSIVENESS, COMPLETENESS, QUALITY, CURRENCY, ERROR-FREE NATURE,
+ COMPATIBILITY, OR FITNESS FOR ANY PARTICULAR PURPOSE OF THE CONTENT. WE ASSUME NO LIABILITY, AND SHALL NOT BE LIABLE FOR,
+ ANY DAMAGES TO, OR VIRUSES OR OTHER MALWARE THAT MAY AFFECT, YOUR COMPUTER EQUIPMENT OR OTHER PROPERTY AS A RESULT OF
+ YOUR DOWNLOADING OF, AND USE OF, ANY CONTENT.
+
+ Limitation of Liability
+ TO THE FULLEST EXTENT PERMITTED UNDER APPLICABLE LAW, IN NO EVENT WILL WE, OR ANY OF OUR EMPLOYEES, AGENTS, OFFICERS, OR
+ TRUSTEES, BE LIABLE FOR DAMAGES OF ANY KIND, UNDER ANY LEGAL THEORY, ARISING OUT OF OR IN CONNECTION WITH YOUR USE, OR
+ INABILITY TO USE, THE CONTENT, INCLUDING ANY DIRECT, INDIRECT, SPECIAL, INCIDENTAL, CONSEQUENTIAL OR PUNITIVE DAMAGES,
+ INCLUDING BUT NOT LIMITED TO, LOSS OF REVENUE, LOSS OF PROFITS, LOSS OF BUSINESS OR ANTICIPATED SAVINGS, LOSS OF USE,
+ LOSS OF GOODWILL, LOSS OF DATA, AND WHETHER CAUSED BY TORT (INCLUDING NEGLIGENCE), BREACH OF CONTRACT OR OTHERWISE,
+ EVEN IF FORESEEABLE. BECAUSE SOME JURISDICTIONS DO NOT ALLOW THE EXCLUSION OR LIMITATION OF LIABLITY FOR CONSEQUENTIAL
+ OR INCIDENTAL DAMAGES, ALL OR A PORTION OF THE ABOVE LIMITATION MAY NOT APPLY TO YOU.
+
+ General
+ If any provision of these Terms and Conditions is held to be invalid, illegal, or unenforceable, then such provision
+ shall be eliminated or limited to the minimum extent such that the remaining provisions of the Terms and Conditions
+ will continue in full force and effect. All matters relating to and arising from the Content or these Terms and Conditions
+ shall be governed by and construed in accordance with the internal laws of the State of Maryland without giving effect
+ to any choice or conflict of law provision or rule. If you choose to download or access the Content from locations
+ outside the United States, you do so at your own risk and you are responsible for compliance with any local laws.
+
+/*********************************************************************/
+
+/*********************************************************************/
+ii. TissueNet dataset
+
+ https://datasets.deepcell.org/
+
+ Modified Apache License
+ Version 2.0, January 2004
+
+ TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
+
+ 1. Definitions.
+
+ "License" shall mean the terms and conditions for use, reproduction,
+ and distribution as defined by Sections 1 through 9 of this document.
+
+ "Licensor" shall mean the copyright owner or entity authorized by
+ the copyright owner that is granting the License.
+
+ "Legal Entity" shall mean the union of the acting entity and all
+ other entities that control, are controlled by, or are under common
+ control with that entity. For the purposes of this definition,
+ "control" means (i) the power, direct or indirect, to cause the
+ direction or management of such entity, whether by contract or
+ otherwise, or (ii) ownership of fifty percent (50%) or more of the
+ outstanding shares, or (iii) beneficial ownership of such entity.
+
+ "You" (or "Your") shall mean an individual or Legal Entity
+ exercising permissions granted by this License.
+
+ "Source" form shall mean the preferred form for making modifications,
+ including but not limited to software source code, documentation
+ source, and configuration files.
+
+ "Object" form shall mean any form resulting from mechanical
+ transformation or translation of a Source form, including but
+ not limited to compiled object code, generated documentation,
+ and conversions to other media types.
+
+ "Work" shall mean the work of authorship, whether in Source or
+ Object form, made available under the License, as indicated by a
+ copyright notice that is included in or attached to the work
+ (an example is provided in the Appendix below).
+
+ "Derivative Works" shall mean any work, whether in Source or Object
+ form, that is based on (or derived from) the Work and for which the
+ editorial revisions, annotations, elaborations, or other modifications
+ represent, as a whole, an original work of authorship. For the purposes
+ of this License, Derivative Works shall not include works that remain
+ separable from, or merely link (or bind by name) to the interfaces of,
+ the Work and Derivative Works thereof.
+
+ "Contribution" shall mean any work of authorship, including
+ the original version of the Work and any modifications or additions
+ to that Work or Derivative Works thereof, that is intentionally
+ submitted to Licensor for inclusion in the Work by the copyright owner
+ or by an individual or Legal Entity authorized to submit on behalf of
+ the copyright owner. For the purposes of this definition, "submitted"
+ means any form of electronic, verbal, or written communication sent
+ to the Licensor or its representatives, including but not limited to
+ communication on electronic mailing lists, source code control systems,
+ and issue tracking systems that are managed by, or on behalf of, the
+ Licensor for the purpose of discussing and improving the Work, but
+ excluding communication that is conspicuously marked or otherwise
+ designated in writing by the copyright owner as "Not a Contribution."
+
+ "Contributor" shall mean Licensor and any individual or Legal Entity
+ on behalf of whom a Contribution has been received by Licensor and
+ subsequently incorporated within the Work.
+
+ 2. Grant of Copyright License. Subject to the terms and conditions of
+ this License, each Contributor hereby grants to You a non-commercial,
+ academic perpetual, worldwide, non-exclusive, no-charge, royalty-free,
+ irrevocable copyright license to reproduce, prepare Derivative Works
+ of, publicly display, publicly perform, sublicense, and distribute the
+ Work and such Derivative Works in Source or Object form. For any other
+ use, including commercial use, please contact: vanvalenlab@gmail.com.
+
+ 3. Grant of Patent License. Subject to the terms and conditions of
+ this License, each Contributor hereby grants to You a non-commercial,
+ academic perpetual, worldwide, non-exclusive, no-charge, royalty-free,
+ irrevocable (except as stated in this section) patent license to make,
+ have made, use, offer to sell, sell, import, and otherwise transfer the
+ Work, where such license applies only to those patent claims licensable
+ by such Contributor that are necessarily infringed by their
+ Contribution(s) alone or by combination of their Contribution(s)
+ with the Work to which such Contribution(s) was submitted. If You
+ institute patent litigation against any entity (including a
+ cross-claim or counterclaim in a lawsuit) alleging that the Work
+ or a Contribution incorporated within the Work constitutes direct
+ or contributory patent infringement, then any patent licenses
+ granted to You under this License for that Work shall terminate
+ as of the date such litigation is filed.
+
+ 4. Redistribution. You may reproduce and distribute copies of the
+ Work or Derivative Works thereof in any medium, with or without
+ modifications, and in Source or Object form, provided that You
+ meet the following conditions:
+
+ (a) You must give any other recipients of the Work or
+ Derivative Works a copy of this License; and
+
+ (b) You must cause any modified files to carry prominent notices
+ stating that You changed the files; and
+
+ (c) You must retain, in the Source form of any Derivative Works
+ that You distribute, all copyright, patent, trademark, and
+ attribution notices from the Source form of the Work,
+ excluding those notices that do not pertain to any part of
+ the Derivative Works; and
+
+ (d) If the Work includes a "NOTICE" text file as part of its
+ distribution, then any Derivative Works that You distribute must
+ include a readable copy of the attribution notices contained
+ within such NOTICE file, excluding those notices that do not
+ pertain to any part of the Derivative Works, in at least one
+ of the following places: within a NOTICE text file distributed
+ as part of the Derivative Works; within the Source form or
+ documentation, if provided along with the Derivative Works; or,
+ within a display generated by the Derivative Works, if and
+ wherever such third-party notices normally appear. The contents
+ of the NOTICE file are for informational purposes only and
+ do not modify the License. You may add Your own attribution
+ notices within Derivative Works that You distribute, alongside
+ or as an addendum to the NOTICE text from the Work, provided
+ that such additional attribution notices cannot be construed
+ as modifying the License.
+
+ You may add Your own copyright statement to Your modifications and
+ may provide additional or different license terms and conditions
+ for use, reproduction, or distribution of Your modifications, or
+ for any such Derivative Works as a whole, provided Your use,
+ reproduction, and distribution of the Work otherwise complies with
+ the conditions stated in this License.
+
+ 5. Submission of Contributions. Unless You explicitly state otherwise,
+ any Contribution intentionally submitted for inclusion in the Work
+ by You to the Licensor shall be under the terms and conditions of
+ this License, without any additional terms or conditions.
+ Notwithstanding the above, nothing herein shall supersede or modify
+ the terms of any separate license agreement you may have executed
+ with Licensor regarding such Contributions.
+
+ 6. Trademarks. This License does not grant permission to use the trade
+ names, trademarks, service marks, or product names of the Licensor,
+ except as required for reasonable and customary use in describing the
+ origin of the Work and reproducing the content of the NOTICE file.
+
+ 7. Disclaimer of Warranty. Unless required by applicable law or
+ agreed to in writing, Licensor provides the Work (and each
+ Contributor provides its Contributions) on an "AS IS" BASIS,
+ WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
+ implied, including, without limitation, any warranties or conditions
+ of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
+ PARTICULAR PURPOSE. You are solely responsible for determining the
+ appropriateness of using or redistributing the Work and assume any
+ risks associated with Your exercise of permissions under this License.
+
+ 8. Limitation of Liability. In no event and under no legal theory,
+ whether in tort (including negligence), contract, or otherwise,
+ unless required by applicable law (such as deliberate and grossly
+ negligent acts) or agreed to in writing, shall any Contributor be
+ liable to You for damages, including any direct, indirect, special,
+ incidental, or consequential damages of any character arising as a
+ result of this License or out of the use or inability to use the
+ Work (including but not limited to damages for loss of goodwill,
+ work stoppage, computer failure or malfunction, or any and all
+ other commercial damages or losses), even if such Contributor
+ has been advised of the possibility of such damages.
+
+ 9. Accepting Warranty or Additional Liability. While redistributing
+ the Work or Derivative Works thereof, You may choose to offer,
+ and charge a fee for, acceptance of support, warranty, indemnity,
+ or other liability obligations and/or rights consistent with this
+ License. However, in accepting such obligations, You may act only
+ on Your own behalf and on Your sole responsibility, not on behalf
+ of any other Contributor, and only if You agree to indemnify,
+ defend, and hold each Contributor harmless for any liability
+ incurred by, or claims asserted against, such Contributor by reason
+ of your accepting any such warranty or additional liability.
+
+ 10. Neither the name of Caltech nor the names of its contributors may be
+ used to endorse or promote products derived from this software without
+ specific prior written permission.
+
+ END OF TERMS AND CONDITIONS
+
+ APPENDIX: How to apply the Apache License to your work.
+
+ To apply the Apache License to your work, attach the following
+ boilerplate notice, with the fields enclosed by brackets "[]"
+ replaced with your own identifying information. (Don't include
+ the brackets!) The text should be enclosed in the appropriate
+ comment syntax for the file format. We also recommend that a
+ file or class name and description of purpose be included on the
+ same "printed page" as the copyright notice for easier
+ identification within third-party archives.
+
+ Copyright [yyyy] [name of copyright owner]
+
+ Licensed under the Apache License, Version 2.0 (the "License");
+ you may not use this file except in compliance with the License.
+ You may obtain a copy of the License at
+
+ http://www.apache.org/licenses/LICENSE-2.0
+
+ Unless required by applicable law or agreed to in writing, software
+ distributed under the License is distributed on an "AS IS" BASIS,
+ WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+ See the License for the specific language governing permissions and
+ limitations under the License.
+
+/*********************************************************************/
+
+/*********************************************************************/
+iii. Kaggle Nuclei Segmentation
+ https://www.nature.com/articles/s41592-019-0612-7#rightslink
+
+ CC BY 4.0
+ http://creativecommons.org/licenses/by/4.0/
+
+/*********************************************************************/
+
+/*********************************************************************/
+iv. Omnipose
+
+ https://github.com/kevinjohncutler/omnipose/blob/main/LICENSE
+
+ Omnipose NonCommercial License
+ Copyright (c) 2021 University of Washington.
+
+ Redistribution and use for noncommercial purposes in source and binary forms, with or without modification, are permitted
+ provided that the following conditions are met:
+ 1. The software is used solely for noncommercial purposes. For commercial use rights, contact University of Washington,
+ CoMotion, at license@uw.edu.
+ 2. Redistributions of source code must retain the above copyright notice, this list of conditions and the below
+ disclaimer.
+ 3. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following
+ disclaimer in the documentation and/or other materials provided with the distribution.
+ 4. Redistributions, with or without modifications, shall only be licensed under this NonCommercial License.
+ 5. Neither the name of the University of Washington nor the names of its contributors may be used to endorse or promote
+ products derived from this software without specific prior written permission.
+
+ THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES,
+ INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
+ DISCLAIMED. IN NO EVENT SHALL THE UNIVERSITY OF WASHINGTON OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT,
+ INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS
+ OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY,
+ WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF
+ THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
+
+/*********************************************************************/
+
+/*********************************************************************/
+v. NIPS Cell Segmentation Challenge
+
+ https://neurips22-cellseg.grand-challenge.org/dataset/
+
+ CC BY-NC-ND
+ https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en
+
+/*********************************************************************/
+
+/*********************************************************************/
+vi. LiveCell
+
+ https://sartorius-research.github.io/LIVECell/
+
+ CC BY-NC 4.0
+ https://creativecommons.org/licenses/by-nc/4.0/
+
+/*********************************************************************/
+
+/*********************************************************************/
+vii. Deepbacs
+
+ https://github.com/HenriquesLab/DeepBacs/blob/main/LICENSE
+
+ CC0 1.0
+ https://creativecommons.org/publicdomain/zero/1.0/deed.en
+
+/*********************************************************************/
+Data Usage Agreement / Citations
diff --git a/models/vista2d/download_preprocessor/all_file_downloader.py b/models/vista2d/download_preprocessor/all_file_downloader.py
new file mode 100644
index 00000000..c35a414e
--- /dev/null
+++ b/models/vista2d/download_preprocessor/all_file_downloader.py
@@ -0,0 +1,80 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import argparse
+import os
+
+import requests
+from tqdm import tqdm
+
+
+def download_files(url_dict, directory):
+ if not os.path.exists(directory):
+ os.makedirs(directory)
+
+ for key, url in url_dict.items():
+ if key == "nips_train.zip" or key == "nips_test.zip":
+ if not os.path.exists(os.path.join(directory, "nips_dataset")):
+ os.mkdir(os.path.join(directory, "nips_dataset"))
+ base_dir = os.path.join(directory, "nips_dataset")
+ elif key == "deepbacs.zip":
+ if not os.path.exists(os.path.join(directory, "deepbacs_dataset")):
+ os.mkdir(os.path.join(directory, "deepbacs_dataset"))
+ base_dir = os.path.join(directory, "deepbacs_dataset")
+ elif key == "livecell":
+ if not os.path.exists(os.path.join(directory, "livecell_dataset")):
+ os.mkdir(os.path.join(directory, "livecell_dataset"))
+ base_dir = os.path.join(directory, "livecell_dataset")
+ print(f"Downloading from {key}: {url}")
+ os.system(url + base_dir)
+ continue
+
+ try:
+ print(f"Downloading from {key}: {url}")
+ response = requests.get(url, stream=True, allow_redirects=True)
+ total_size = int(response.headers.get("content-length", 0))
+
+ # Extract the filename from the URL or use the key as the filename
+ filename = os.path.basename(key)
+ file_path = os.path.join(base_dir, filename)
+
+ # Write the content to a file in the specified directory with progress
+ with open(file_path, "wb") as file, tqdm(
+ desc=filename, total=total_size, unit="iB", unit_scale=True, unit_divisor=1024
+ ) as bar:
+ for data in response.iter_content(chunk_size=1024):
+ size = file.write(data)
+ bar.update(size)
+
+ print(f"Saved to {file_path}")
+ except Exception as e:
+ print(f"Failed to download from {key} ({url}). Reason: {str(e)}")
+
+
+def main():
+ parser = argparse.ArgumentParser(description="Process some integers.")
+ parser.add_argument("--dir", type=str, help="Directory to download files to", default="/set/the/path")
+
+ args = parser.parse_args()
+ directory = os.path.normpath(args.dir)
+
+ url_dict = {
+ "deepbacs.zip": "https://zenodo.org/records/5551009/files/DeepBacs_Data_Segmentation_StarDist_MIXED_dataset.zip?download=1",
+ "nips_test.zip": "https://zenodo.org/records/10719375/files/Testing.zip?download=1",
+ "nips_train.zip": "https://zenodo.org/records/10719375/files/Training-labeled.zip?download=1",
+ "livecell": "wget --recursive --no-parent --cut-dirs=0 --timestamping -i urls.txt --directory-prefix=",
+ # Add URLs with keys here
+ }
+ download_files(url_dict, directory)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/models/vista2d/download_preprocessor/cellpose_agreement.png b/models/vista2d/download_preprocessor/cellpose_agreement.png
new file mode 100644
index 00000000..79e09aa0
Binary files /dev/null and b/models/vista2d/download_preprocessor/cellpose_agreement.png differ
diff --git a/models/vista2d/download_preprocessor/cellpose_links.png b/models/vista2d/download_preprocessor/cellpose_links.png
new file mode 100644
index 00000000..ca18b62f
Binary files /dev/null and b/models/vista2d/download_preprocessor/cellpose_links.png differ
diff --git a/models/vista2d/download_preprocessor/data_tree.png b/models/vista2d/download_preprocessor/data_tree.png
new file mode 100644
index 00000000..bff4e5ab
Binary files /dev/null and b/models/vista2d/download_preprocessor/data_tree.png differ
diff --git a/models/vista2d/download_preprocessor/generate_json.py b/models/vista2d/download_preprocessor/generate_json.py
new file mode 100644
index 00000000..0257cb1d
--- /dev/null
+++ b/models/vista2d/download_preprocessor/generate_json.py
@@ -0,0 +1,993 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import argparse
+import gc
+import json
+import os
+import shutil
+import time
+import warnings
+import zipfile
+
+import imageio.v3 as imageio
+import numpy as np
+from PIL import Image
+from pycocotools.coco import COCO
+from sklearn.model_selection import KFold
+
+# from skimage.io import imsave
+# from skimage.measure import label
+# import imageio
+
+
+def min_label_precision(label):
+ lm = label.max()
+
+ if lm <= 255:
+ label = label.astype(np.uint8)
+ elif lm <= 65535:
+ label = label.astype(np.uint16)
+ else:
+ label = label.astype(np.uint32)
+
+ return label
+
+
+def guess_convert_to_uint16(img, margin=30):
+ """
+ Guess a multiplier that makes all pixels integers.
+ The input img (each channel) is already in the range 0..1, they must have been converted from uint16 integers as image / scale,
+ where scale was the unknown max intensity.
+ We could guess the scale by looking at unique values: 1/np.min(np.diff(np.unique(im)).
+ the hypothesis is that it will be more accurate recovery of the original image,
+ instead of doing a simple (img*65535).astype(np.uint16)
+ """
+
+ for i in range(img.shape[0]):
+ im = img[i]
+
+ if im.any():
+ start = time.time()
+ imsmall = im[::4, ::4] # subsample
+ # imsmall = im
+
+ scale = int(np.round(1 / np.min(np.diff(np.unique(imsmall))))) # guessing scale
+ test = [
+ (np.sum((imsmall * k) % 1)) for k in range(scale - margin, scale + margin)
+ ] # finetune, guess a multiplier that makes all pixels integers
+ sid = np.argmin(test) # fine tune scale
+
+ if scale < 16000 or scale > 16400:
+ warnings.warn("scale not in expected range")
+ print(
+ "guessing scale",
+ scale,
+ test[margin],
+ "fine tuning scale",
+ scale - margin + sid,
+ "dif",
+ test[sid],
+ "time",
+ time.time() - start,
+ )
+
+ scale = 16384
+ else:
+ scale = scale - margin + sid
+ # all the recovered scale values seems to be up to 16384,
+ # we can stretch to 65535(for better visualization, most tiff viewers expect that range)
+ scale = min(65535, scale * 4)
+ img[i] = im * scale
+
+ img = img.astype(np.uint16)
+ return img
+
+
+def concatenate_masks(mask_dir):
+ labeled_mask = None
+ i = 0
+ for filename in sorted(os.listdir(mask_dir)):
+ if filename.endswith(".png"):
+ mask = imageio.imread(os.path.join(mask_dir, filename))
+ if labeled_mask is None:
+ labeled_mask = np.zeros(shape=mask.shape, dtype=np.uint16)
+ labeled_mask[mask > 0] = i
+ i = i + 1
+
+ if i <= 255:
+ labeled_mask = labeled_mask.astype(np.uint8)
+
+ return labeled_mask
+
+
+# def concatenate_masks(mask_dir):
+# masks = []
+# for filename in sorted(os.listdir(mask_dir)):
+# if filename.endswith('.png'):
+# mask = imageio.imread(os.path.join(mask_dir, filename))
+# masks.append(mask)
+# concatenated_mask = np.any(masks, axis=0).astype(np.uint8)
+# labeled_mask = label(concatenated_mask)
+# return labeled_mask
+
+# def normalize_image(image):
+# # Convert to float and normalize each channel
+# image = image.astype(np.float32)
+# for i in range(3):
+# channel = image[..., i]
+# channel_min = np.min(channel)
+# channel_max = np.max(channel)
+# if channel_max - channel_min != 0:
+# image[..., i] = (channel - channel_min) / (channel_max - channel_min)
+# return image
+
+
+def get_filenames_exclude_masks(dir1, target_string):
+ filenames = []
+ # Combine lists of files from both directories
+ files = os.listdir(dir1)
+ # Filter files that contain the target string but exclude 'masks'
+ filenames = [f for f in files if target_string in f and "masks" not in f]
+
+ return filenames
+
+
+def remove_overlaps(masks, medians, overlap_threshold=0.75):
+ """replace overlapping mask pixels with mask id of closest mask
+ if mask fully within another mask, remove it
+ masks = Nmasks x Ly x Lx
+ """
+ cellpix = masks.sum(axis=0)
+ igood = np.ones(masks.shape[0], "bool")
+ for i in masks.sum(axis=(1, 2)).argsort():
+ npix = float(masks[i].sum())
+ noverlap = float(masks[i][cellpix > 1].sum())
+ if noverlap / npix >= overlap_threshold:
+ igood[i] = False
+ cellpix[masks[i] > 0] -= 1
+ # print(cellpix.min())
+ print(f"removing {(~igood).sum()} masks")
+ masks = masks[igood]
+ medians = medians[igood]
+ cellpix = masks.sum(axis=0)
+ overlaps = np.array(np.nonzero(cellpix > 1.0)).T
+ dists = ((overlaps[:, :, np.newaxis] - medians.T) ** 2).sum(axis=1)
+ tocell = np.argmin(dists, axis=1)
+ masks[:, overlaps[:, 0], overlaps[:, 1]] = 0
+ masks[tocell, overlaps[:, 0], overlaps[:, 1]] = 1
+
+ # labels should be 1 to mask.shape[0]
+ masks = masks.astype(int) * np.arange(1, masks.shape[0] + 1, 1, int)[:, np.newaxis, np.newaxis]
+ masks = masks.sum(axis=0)
+ gc.collect()
+ return masks
+
+
+def livecell_json_files(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of livecell extracted dataset as input and
+ creates 7 json lists with 5 folds. Separate testing set is recorded in the json list.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ At the time of creation, the livecell zipfile had 'images' and 'LIVECell_dataset_2021' directories
+ """
+
+ # "A172", "BT474", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"
+ # TODO "BV2" is being skipped
+ cell_type_list = ["A172", "BT474", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"]
+ for each_cell_tp in cell_type_list:
+ for split in ["train", "val", "test"]:
+ print(f"Working on split: {split}")
+
+ if split == "test":
+ img_path = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp)
+ msk_path = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp + "_masks")
+ else:
+ img_path = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp)
+ msk_path = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp + "_masks")
+ if not os.path.exists(msk_path):
+ os.makedirs(msk_path)
+
+ # annotation path
+ path = os.path.join(
+ dataset_dir,
+ "livecell-dataset.s3.eu-central-1.amazonaws.com",
+ "LIVECell_dataset_2021",
+ "annotations",
+ "LIVECell_single_cells",
+ each_cell_tp.lower(),
+ split + ".json",
+ )
+ annotation = COCO(path)
+ # Convert COCO format segmentation to binary mask
+ images = annotation.loadImgs(annotation.getImgIds())
+ height = []
+ width = []
+ for index, im in enumerate(images):
+ print("Status: {}/{}, Process image: {}".format(index, len(images), im["file_name"]))
+ if (
+ im["file_name"] == "BV2_Phase_C4_2_03d00h00m_1.tif"
+ or im["file_name"] == "BV2_Phase_C4_2_03d00h00m_3.tif"
+ ):
+ print("Skipping the file: BV2_Phase_C4_2_03d00h00m_1.tif, as it is troublesome")
+ continue
+ # load image
+ img = Image.open(os.path.join(img_path, im["file_name"])).convert("L")
+ height.append(img.size[0])
+ width.append(img.size[1])
+ # arr = np.asarray(img) #? not used
+ # msk = np.zeros(arr.shape)
+ # load and display instance annotations
+ annids = annotation.getAnnIds(imgIds=im["id"], iscrowd=None)
+ anns = annotation.loadAnns(annids)
+ idx = 1
+ medians = []
+ masks = []
+ k = 0
+ for ann in anns:
+ # convert segmentation to binary mask
+ mask = annotation.annToMask(ann)
+ masks.append(mask)
+ ypix, xpix = mask.nonzero()
+ medians.append(np.array([ypix.mean().astype(np.float32), xpix.mean().astype(np.float32)]))
+ k += 1
+ # add instance mask to image mask
+ # msk = np.add(msk, mask*idx)
+ # idx += 1
+
+ masks = np.array(masks).astype(np.int8)
+ medians = np.array(medians)
+ masks = remove_overlaps(masks, medians, overlap_threshold=0.75)
+ gc.collect()
+
+ # ## Create new name for the image and also for the mask and save them as .tif format
+ # masks_int32 = masks.astype(np.int32)
+ # mask_pil = Image.fromarray(masks_int32, 'I')
+
+ t_filename = im["file_name"]
+ # cell_type = t_filename.split('_')[0] #? not used
+ new_mask_name = t_filename[:-4] + "_masks.tif"
+ # mask_pil.save(os.path.join(msk_path, new_mask_name))
+ imageio.imwrite(os.path.join(msk_path, new_mask_name), min_label_precision(masks))
+ gc.collect()
+
+ print(f"In total {len(images)} images")
+
+ # The directory containing your files
+ # cell_type = 'BV2'
+ json_save_path = os.path.join(json_f_path, f"lc_{each_cell_tp}.json")
+ directory = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp)
+ mask_directory = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp + "_masks")
+ test_directory = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp)
+ mask_test_directory = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp + "_masks")
+ # List to hold all image-mask pairs
+ data_pairs = []
+ test_data_pairs = []
+ all_data = {}
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(directory):
+ if filename.endswith(".tif"):
+ # Construct the corresponding mask filename
+ mask_filename = filename.replace(".tif", "_masks.tif")
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(mask_directory, mask_filename)):
+ # Add the pair to the list
+ data_pairs.append(
+ {
+ "image": os.path.join(
+ "livecell_dataset", "images", "livecell_train_val_images", each_cell_tp, filename
+ ),
+ "label": os.path.join(
+ "livecell_dataset",
+ "images",
+ "livecell_train_val_images",
+ f"{each_cell_tp}_masks",
+ mask_filename,
+ ),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(data_pairs)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(test_directory):
+ if filename.endswith(".tif"):
+ # Construct the corresponding mask filename
+ mask_filename = filename.replace(".tif", "_masks.tif")
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(mask_test_directory, mask_filename)):
+ # Add the pair to the list
+ test_data_pairs.append(
+ {
+ "image": os.path.join(
+ "livecell_dataset", "images", "livecell_test_images", each_cell_tp, filename
+ ),
+ "label": os.path.join(
+ "livecell_dataset",
+ "images",
+ "livecell_test_images",
+ f"{each_cell_tp}_masks",
+ mask_filename,
+ ),
+ }
+ )
+
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = test_data_pairs
+
+ with open(json_save_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def tissuenet_json_files(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of TissueNet extracted dataset as input and
+ creates 13 json lists with 5 folds each. Separate testing set is recorded in the json list per subset.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ At the time of creation, the tissuenet 1.0 zipfile had 'train', 'val' and 'test' directories that
+ images with paired labels.
+ """
+
+ for folder in ["train", "val", "test"]:
+ if not os.path.exists(os.path.join(dataset_dir, "tissuenet_1.0", folder)):
+ os.mkdir(os.path.join(dataset_dir, "tissuenet_1.0", folder))
+
+ for folder in ["train", "val", "test"]:
+ print(f"Working on {folder} directory of tissuenet")
+ f_name = f"tissuenet_1.0/tissuenet_v1.0_{folder}.npz"
+ dat = np.load(os.path.join(dataset_dir, f_name))
+ data = dat["X"]
+ labels = dat["y"]
+ tissues = dat["tissue_list"]
+ platforms = dat["platform_list"]
+ tlabels = np.unique(tissues)
+ plabels = np.unique(platforms)
+ tp = 0
+ for t in tlabels:
+ for p in plabels:
+ ix = ((tissues == t) * (platforms == p)).nonzero()[0]
+ tp += 1
+ if len(ix) > 0:
+ print(f"Working on {t} {p}")
+
+ for k, i in enumerate(ix):
+ print(f"Status: {k}/{len(ix)} {tp}/{len(tlabels) * len(plabels)} {t} {p}")
+ img = data[i].transpose(2, 0, 1)
+ label = labels[i][:, :, 0]
+
+ img = guess_convert_to_uint16(img) # guess inverse scale and convert to uint16
+ label = min_label_precision(label)
+
+ if folder == "train":
+ img = img.reshape(2, 2, 256, 2, 256).transpose(0, 1, 3, 2, 4).reshape(2, 4, 256, 256)
+ label = label.reshape(2, 256, 2, 256).transpose(0, 2, 1, 3).reshape(4, 256, 256)
+
+ zero_channel = np.zeros((1, img.shape[1], img.shape[2], img.shape[3]), dtype=img.dtype)
+
+ # Concatenate the zero channel with the original array along the first dimension
+ new_array = np.concatenate([img, zero_channel], axis=0)
+ # reshaped_array = np.transpose(new_array, (1, 2, 3, 0))
+ for j in range(4):
+ img_name = f"{folder}/{t}_{p}_{k}_{j}.tif"
+ mask_name = f"{folder}/{t}_{p}_{k}_{j}_masks.tif"
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", img_name), new_array[:, j])
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", mask_name), label[j])
+ else:
+ zero_channel = np.zeros((1, img.shape[1], img.shape[2]), dtype=img.dtype)
+ new_array = np.concatenate([img, zero_channel], axis=0)
+ # reshaped_array = np.transpose(new_array, (1, 2, 0))
+ img_name = f"{folder}/{t}_{p}_{k}.tif"
+ mask_name = f"{folder}/{t}_{p}_{k}_masks.tif"
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", img_name), new_array)
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", mask_name), label)
+
+ t_p_combos = [
+ ["breast", "imc"],
+ ["breast", "mibi"],
+ ["breast", "vectra"],
+ ["gi", "codex"],
+ ["gi", "mibi"],
+ ["gi", "mxif"],
+ ["immune", "cycif"],
+ ["immune", "mibi"],
+ ["immune", "vectra"],
+ ["lung", "cycif"],
+ ["lung", "mibi"],
+ ["pancreas", "codex"],
+ ["pancreas", "vectra"],
+ ["skin", "mibi"],
+ ]
+
+ for each_t_p in t_p_combos:
+ json_f_name = "tn_" + each_t_p[0] + "_" + each_t_p[1] + ".json"
+ json_f_subset_path = os.path.join(json_f_path, json_f_name)
+
+ tp_match = each_t_p[0] + "_" + each_t_p[1]
+ train_filenames = get_filenames_exclude_masks(os.path.join(dataset_dir, "tissuenet_1.0", "train"), tp_match)
+ val_filenames = get_filenames_exclude_masks(os.path.join(dataset_dir, "tissuenet_1.0", "val"), tp_match)
+ test_filenames = get_filenames_exclude_masks(os.path.join(dataset_dir, "tissuenet_1.0", "test"), tp_match)
+
+ train_data_list = []
+ test_data_list = []
+
+ for each_tf in train_filenames:
+ t_dict = {
+ "image": os.path.join("tissuenet_dataset", "tissuenet_1.0", "train", each_tf),
+ "label": os.path.join("tissuenet_dataset", "tissuenet_1.0", "train", each_tf[:-4] + "_masks.tif"),
+ }
+ train_data_list.append(t_dict)
+
+ for each_vf in val_filenames:
+ t_dict = {
+ "image": os.path.join("tissuenet_dataset", "tissuenet_1.0", "val", each_vf),
+ "label": os.path.join("tissuenet_dataset", "tissuenet_1.0", "val", each_vf[:-4] + "_masks.tif"),
+ }
+ train_data_list.append(t_dict)
+
+ for each_tf in test_filenames:
+ t_dict = {
+ "image": os.path.join("tissuenet_dataset", "tissuenet_1.0", "test", each_tf),
+ "label": os.path.join("tissuenet_dataset", "tissuenet_1.0", "test", each_tf[:-4] + "_masks.tif"),
+ }
+ test_data_list.append(t_dict)
+
+ # print(train_data_list)
+ # print(test_data_list)
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(train_data_list)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ all_data = {}
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = test_data_list
+
+ with open(json_f_subset_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def omnipose_json_file(dataset_dir, json_path):
+ """
+ This function takes in the directory of extracted Omnipose dataset as input
+ and creates a json list with 5 folds. Please note that only 'bact_phase' and 'bact_fluor' were
+ used for creating datasets as they have bacteria the other directiories are worms. Each directory
+ has 'train_sorted' and 'test_sorted'.Separate testing set is recorded in the json list.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ """
+ # Define the folders
+ op_list = ["bact_fluor", "bact_phase"]
+ for each_op in op_list:
+ print(f"Working on {each_op} ...")
+ images_folder = os.path.join(dataset_dir, each_op, "train_sorted")
+ test_images_folder = os.path.join(dataset_dir, each_op, "test_sorted")
+ json_f_path = os.path.join(json_path, f"op_{each_op}.json")
+
+ # Initialize the list for training data
+ training_data = []
+
+ # Loop through each image file to find its corresponding label file
+ sub_dirs = os.listdir(images_folder)
+ # Likely Omnipose dataset was created using a Mac and hence the spare filename
+ sub_dirs.remove(".DS_Store")
+ for each_sub in sub_dirs:
+ # List files in the images folder
+ image_files = os.listdir(os.path.join(images_folder, each_sub))
+ for image_file in image_files:
+ # Extract the name without the extension
+ base_name = os.path.splitext(image_file)[0]
+
+ # Construct the label file name by adding '_label' before the extension
+ label_file = base_name + "_masks.tif" # + os.path.splitext(image_file)[1]
+ flows_file = base_name + "_flows.tif"
+ # Check if the corresponding label file exists in the labels folder
+ if label_file in os.listdir(os.path.join(images_folder, each_sub)):
+ # Add the file names to the training data list
+ training_data.append(
+ {
+ "image": os.path.join("omnipose_dataset", each_op, "train_sorted", each_sub, image_file),
+ "label": os.path.join("omnipose_dataset", each_op, "train_sorted", each_sub, label_file),
+ "flows": os.path.join("omnipose_dataset", each_op, "train_sorted", each_sub, flows_file),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(training_data)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ # Initialize the list for testing data
+ testing_data = []
+
+ test_sub_dirs = os.listdir(test_images_folder)
+ # Likely Omnipose dataset was created using a Mac and hence the spare filename
+ test_sub_dirs.remove(".DS_Store")
+ # Loop through each image file to find its corresponding label file
+ for each_test_sub in test_sub_dirs:
+ # List files in the images folder
+ test_image_files = os.listdir(os.path.join(test_images_folder, each_test_sub))
+ for image_file in test_image_files:
+ # Extract the name without the extension
+ base_name = os.path.splitext(image_file)[0]
+
+ # Construct the label file name by adding '_label' before the extension
+ label_file = base_name + "_masks.tif" # + os.path.splitext(image_file)[1]
+
+ # Check if the corresponding label file exists in the labels folder
+ if label_file in os.listdir(os.path.join(test_images_folder, each_test_sub)):
+ # Add the file names to the training data list
+ testing_data.append(
+ {
+ "image": os.path.join(
+ "omnipose_dataset", each_op, "test_sorted", each_test_sub, image_file
+ ),
+ "label": os.path.join(
+ "omnipose_dataset", each_op, "test_sorted", each_test_sub, label_file
+ ),
+ }
+ )
+
+ all_data = {}
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = testing_data
+
+ # Save the training data list to a JSON file
+ with open(json_f_path, "w") as json_file:
+ json.dump(all_data, json_file, indent=4)
+
+
+def nips_json_file(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of extracted NIPS cell segmentation challenge as input
+ and creates a json list with 5 folds. Separate testing set is recorded in the json list.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ At the time of creation, the NIPS zipfile had 'Training-labeled' and 'Testing' directories that
+ both contained 'images' and 'labels' directories
+ """
+ # The directory containing your files
+ json_save_path = os.path.normpath(json_f_path)
+ directory = os.path.join(dataset_dir, "Training-labeled")
+ test_directory = os.path.join(dataset_dir, "Testing", "Public")
+ # List to hold all image-mask pairs
+ data_pairs = []
+ test_data_pairs = []
+ all_data = {}
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(os.path.join(directory, "images")):
+ if os.path.exists(os.path.join(directory, "images", filename)):
+ # Extract the name without the extension
+ base_name = os.path.splitext(filename)[0]
+
+ # Construct the label file name by adding '_label' before the extension
+ label_file = base_name + "_label.tiff" # + os.path.splitext(image_file)[1]
+
+ # Check if the corresponding label file exists in the labels folder
+ if label_file in os.listdir(os.path.join(directory, "labels")):
+ # Add the file names to the training data list
+ data_pairs.append(
+ {
+ "image": os.path.join("nips_dataset", "Training-labeled", "images", filename),
+ "label": os.path.join("nips_dataset", "Training-labeled", "labels", label_file),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(data_pairs)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(os.path.join(test_directory, "images")):
+ if os.path.exists(os.path.join(test_directory, "images", filename)):
+ # Extract the name without the extension
+ base_name = os.path.splitext(filename)[0]
+
+ # Construct the label file name by adding '_label' before the extension
+ label_file = base_name + "_label.tiff" # + os.path.splitext(image_file)[1]
+
+ # Check if the corresponding label file exists in the labels folder
+ if label_file in os.listdir(os.path.join(test_directory, "labels")):
+ # Add the file names to the training data list
+ test_data_pairs.append(
+ {
+ "image": os.path.join("nips_dataset", "Testing", "Public", "images", filename),
+ "label": os.path.join("nips_dataset", "Testing", "Public", "labels", label_file),
+ }
+ )
+
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = test_data_pairs
+
+ with open(json_save_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def kaggle_json_file(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of kaggle nuclei extracted dataset as input and
+ creates a json list with 5 folds.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ The function creates an instance processed dataset and then a 5 fold json file based on
+ the instance processed dataset
+ """
+ data_dir = os.path.join(dataset_dir, "stage1_train")
+ saving_path = os.path.join(dataset_dir, "instance_processed_data")
+ if not os.path.exists(saving_path):
+ os.mkdir(saving_path)
+
+ # Process the images and create instance masks first
+ for idx, subdir in enumerate(os.listdir(data_dir)):
+ subdir_path = os.path.join(data_dir, subdir)
+ if os.path.isdir(subdir_path):
+ images_dir = os.path.join(subdir_path, "images")
+ masks_dir = os.path.join(subdir_path, "masks")
+ if os.path.isdir(images_dir) and os.path.isdir(masks_dir):
+ image_file = os.path.join(images_dir, os.listdir(images_dir)[0])
+ filename_prefix = f"kg_bowl_{idx}_"
+
+ mask_data = concatenate_masks(masks_dir)
+
+ # ## Apply channel-wise normalization and use only the first three channels
+ # image_data = imageio.imread(image_file)
+ # normalized_image = normalize_image(image_data[..., :3])
+ # imageio.imwrite(os.path.join(saving_path, f"{filename_prefix}img.tiff"), normalized_image)
+ shutil.copyfile(image_file, os.path.join(saving_path, f"{filename_prefix}img.png"))
+ imageio.imwrite(os.path.join(saving_path, f"{filename_prefix}img_masks.tiff"), mask_data)
+
+ directory = saving_path
+
+ # List to hold all image-mask pairs
+ data_pairs = []
+ all_data = {}
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(directory):
+ if filename.endswith("_img.png"):
+ # Construct the corresponding mask filename
+ mask_filename = filename.replace("_img.png", "_img_masks.tiff")
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(directory, mask_filename)):
+ # Add the pair to the list
+ data_pairs.append(
+ {
+ "image": os.path.join("kaggle_dataset", "instance_processed_data", filename),
+ "label": os.path.join("kaggle_dataset", "instance_processed_data", mask_filename),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(data_pairs)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ all_data["training"] = sorted_data_pairs
+
+ with open(json_f_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def deepbacs_json_file(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of deepbacs extracted dataset as input and
+ creates a json list with 5 folds. Separate testing set is recorded in the json list.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ At the time of creation, the deepbacs zipfile had 'training' and 'test' directories that
+ both contained 'source' and 'target' directories
+ """
+ # The directory containing your files
+ json_save_path = os.path.normpath(json_f_path)
+ directory = os.path.join(dataset_dir, "training")
+ test_directory = os.path.join(dataset_dir, "test")
+ # List to hold all image-mask pairs
+ data_pairs = []
+ test_data_pairs = []
+ all_data = {}
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(os.path.join(directory, "source")):
+ if os.path.exists(os.path.join(directory, "source", filename)):
+ # Construct the corresponding mask filename
+ mask_filename = filename
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(directory, "target", mask_filename)):
+ # Add the pair to the list
+ data_pairs.append(
+ {
+ "image": os.path.join("deepbacs_dataset", "training", "source", filename),
+ "label": os.path.join("deepbacs_dataset", "training", "target", mask_filename),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(data_pairs)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(os.path.join(test_directory, "source")):
+ if os.path.exists(os.path.join(test_directory, "source", filename)):
+ # Construct the corresponding mask filename
+ mask_filename = filename
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(test_directory, "target", mask_filename)):
+ # Add the pair to the list
+ test_data_pairs.append(
+ {
+ "image": os.path.join("deepbacs_dataset", "test", "source", filename),
+ "label": os.path.join("deepbacs_dataset", "test", "target", filename),
+ }
+ )
+
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = test_data_pairs
+
+ with open(json_save_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def cellpose_json_file(dataset_dir, json_f_path):
+ """
+ This function takes in the directory of cellpose extracted dataset as input and
+ creates a json list with 5 folds. Separate testing set is recorded in the json list.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ At the time of creation, the cellpose dataset had 'train.zip' and 'test.zip' that
+ extracted as 'train' and 'test' directories
+ """
+ # The directory containing your files
+ json_save_path = os.path.normpath(json_f_path)
+ directory = os.path.join(dataset_dir, "train")
+ test_directory = os.path.join(dataset_dir, "test")
+
+ # List to hold all image-mask pairs
+ data_pairs = []
+ test_data_pairs = []
+ all_data = {}
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(directory):
+ if filename.endswith("_img.png"):
+ # Construct the corresponding mask filename
+ mask_filename = filename.replace("_img.png", "_masks.png")
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.normpath(os.path.join(directory, mask_filename))):
+ # Add the pair to the list
+ data_pairs.append(
+ {
+ "image": os.path.join("cellpose_dataset", "train", filename),
+ "label": os.path.join("cellpose_dataset", "train", mask_filename),
+ }
+ )
+
+ # Convert data_pairs to a numpy array for easy indexing by KFold
+ data_pairs_array = np.array(data_pairs)
+
+ # Initialize KFold
+ kf = KFold(n_splits=5, shuffle=True, random_state=42)
+
+ # Assign fold numbers
+ for fold, (_train_index, val_index) in enumerate(kf.split(data_pairs_array)):
+ for idx in val_index:
+ data_pairs_array[idx]["fold"] = fold
+
+ # Convert the array back to a list and sort by fold
+ sorted_data_pairs = sorted(data_pairs_array.tolist(), key=lambda x: x["fold"])
+
+ print(sorted_data_pairs)
+
+ # Scan the directory for image files and create pairs
+ for filename in os.listdir(test_directory):
+ if filename.endswith("_img.png"):
+ # Construct the corresponding mask filename
+ mask_filename = filename.replace("_img.png", "_masks.png")
+
+ # Check if the corresponding mask file exists
+ if os.path.exists(os.path.join(directory, mask_filename)):
+ # Add the pair to the list
+ test_data_pairs.append(
+ {
+ "image": os.path.join("cellpose_dataset", "test", filename),
+ "label": os.path.join("cellpose_dataset", "test", mask_filename),
+ }
+ )
+
+ all_data["training"] = sorted_data_pairs
+ all_data["testing"] = test_data_pairs
+
+ with open(json_save_path, "w") as j_file:
+ json.dump(all_data, j_file, indent=4)
+ j_file.close()
+
+
+def extract_zip(zip_path, extract_to):
+ # Ensure the target directory exists
+ print(f"Extracting from: {zip_path}")
+ print(f"Extracting to: {extract_to}")
+
+ if not os.path.exists(extract_to):
+ os.makedirs(extract_to)
+
+ # Extract all contents of the zip file to the specified directory
+ with zipfile.ZipFile(zip_path, "r") as zip_ref:
+ zip_ref.extractall(extract_to)
+
+
+def main():
+ parser = argparse.ArgumentParser(description="Process some integers.")
+ parser.add_argument("--dir", type=str, help="Directory of datasets to generate json", default="/set/the/path")
+
+ args = parser.parse_args()
+ data_root_path = os.path.normpath(args.dir)
+
+ if not os.path.exists(os.path.join(data_root_path, "json_files")):
+ os.mkdir(os.path.join(data_root_path, "json_files"))
+
+ dataset_dict = {
+ "cellpose_dataset": ["train.zip", "test.zip"],
+ "deepbacs_dataset": ["deepbacs.zip"],
+ "kaggle_dataset": ["data-science-bowl-2018.zip"],
+ "nips_dataset": ["nips_train.zip", "nips_test.zip"],
+ "omnipose_dataset": ["datasets.zip"],
+ "tissuenet_dataset": ["tissuenet_v1.0.zip"],
+ "livecell_dataset": [
+ "livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/images_per_celltype.zip"
+ ],
+ }
+
+ for key, value in dataset_dict.items():
+ dataset_path = os.path.join(data_root_path, key)
+
+ for each_zipped in value:
+ in_path = os.path.join(dataset_path, each_zipped)
+ try:
+ if os.path.exists(in_path):
+ print(f"File exists at: {in_path}")
+ except Exception:
+ print(f"File: {in_path} was not found")
+ out_path = os.path.join(dataset_path)
+ extract_zip(in_path, out_path)
+
+ print(
+ "If we reached here, that means all zip files got extracted ... Working on pre-processing and generating json files"
+ )
+
+ # Looping over all datasets again, Cellpose & Deepbacs have a similar directory structure
+ for key, _value in dataset_dict.items():
+ if key == "cellpose_dataset":
+ print("Creating Cellpose Dataset Json file ...")
+ dataset_path = os.path.join(data_root_path, key)
+ json_path = os.path.join(data_root_path, "json_files", "cellpose.json")
+ cellpose_json_file(dataset_dir=dataset_path, json_f_path=json_path)
+
+ elif key == "nips_dataset":
+ print("Creating NIPS Dataset Json file ...")
+ dataset_path = os.path.join(data_root_path, key)
+ json_path = os.path.join(data_root_path, "json_files", "nips.json")
+ nips_json_file(dataset_dir=dataset_path, json_f_path=json_path)
+
+ elif key == "omnipose_dataset":
+ print("Creating Omnipose Dataset Json files ...")
+ dataset_path = os.path.join(data_root_path, key)
+ json_path = os.path.join(data_root_path, "json_files")
+ omnipose_json_file(dataset_dir=dataset_path, json_path=json_path)
+
+ elif key == "kaggle_dataset":
+ print("Needs additional extraction")
+ train_zip_path = os.path.join(data_root_path, key, "stage1_train.zip")
+ zip_out_path = os.path.join(data_root_path, key, "stage1_train")
+ extract_zip(train_zip_path, zip_out_path)
+ print("Creating Kaggle Dataset Json files ...")
+ dataset_path = os.path.join(data_root_path, key)
+ json_f_path = os.path.join(data_root_path, "json_files", "kaggle.json")
+ kaggle_json_file(dataset_dir=dataset_path, json_f_path=json_f_path)
+
+ elif key == "livecell_dataset":
+ print("Creating LiveCell Dataset Json files ... Please note that 7 files will be created from livecell")
+ dataset_path = os.path.join(data_root_path, key)
+ json_base_name = os.path.join(data_root_path, "json_files")
+ livecell_json_files(dataset_dir=dataset_path, json_f_path=json_base_name)
+
+ elif key == "deepbacs_dataset":
+ print("Creating Deepbacs Dataset Json file ...")
+ dataset_path = os.path.join(data_root_path, key)
+ json_path = os.path.join(data_root_path, "json_files", "deepbacs.json")
+ deepbacs_json_file(dataset_dir=dataset_path, json_f_path=json_path)
+
+ elif key == "tissuenet_dataset":
+ print("Creating TissueNet Dataset Json files ... Please note that 13 files will be created from tissuenet")
+ dataset_path = os.path.join(data_root_path, key)
+ json_base_name = os.path.join(data_root_path, "json_files")
+ tissuenet_json_files(dataset_dir=dataset_path, json_f_path=json_base_name)
+
+ return None
+
+
+if __name__ == "__main__":
+ main()
diff --git a/models/vista2d/download_preprocessor/kaggle_download.png b/models/vista2d/download_preprocessor/kaggle_download.png
new file mode 100644
index 00000000..3639fb8d
Binary files /dev/null and b/models/vista2d/download_preprocessor/kaggle_download.png differ
diff --git a/models/vista2d/download_preprocessor/omnipose_download.png b/models/vista2d/download_preprocessor/omnipose_download.png
new file mode 100644
index 00000000..7a34e1f6
Binary files /dev/null and b/models/vista2d/download_preprocessor/omnipose_download.png differ
diff --git a/models/vista2d/download_preprocessor/process_data.py b/models/vista2d/download_preprocessor/process_data.py
new file mode 100644
index 00000000..45ad7ec2
--- /dev/null
+++ b/models/vista2d/download_preprocessor/process_data.py
@@ -0,0 +1,399 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import argparse
+import gc
+import os
+import shutil
+import time
+import warnings
+import zipfile
+
+import imageio.v3 as imageio
+import numpy as np
+from PIL import Image
+from pycocotools.coco import COCO
+
+
+def min_label_precision(label):
+ lm = label.max()
+
+ if lm <= 255:
+ label = label.astype(np.uint8)
+ elif lm <= 65535:
+ label = label.astype(np.uint16)
+ else:
+ label = label.astype(np.uint32)
+
+ return label
+
+
+def guess_convert_to_uint16(img, margin=30):
+ """
+ Guess a multiplier that makes all pixels integers.
+ The input img (each channel) is already in the range 0..1, they must have been converted from uint16 integers as image / scale,
+ where scale was the unknown max intensity.
+ We could guess the scale by looking at unique values: 1/np.min(np.diff(np.unique(im)).
+ the hypothesis is that it will be more accurate recovery of the original image,
+ instead of doing a simple (img*65535).astype(np.uint16)
+ """
+
+ for i in range(img.shape[0]):
+ im = img[i]
+
+ if im.any():
+ start = time.time()
+ imsmall = im[::4, ::4] # subsample
+ # imsmall = im
+
+ scale = int(np.round(1 / np.min(np.diff(np.unique(imsmall))))) # guessing scale
+ test = [
+ (np.sum((imsmall * k) % 1)) for k in range(scale - margin, scale + margin)
+ ] # finetune, guess a multiplier that makes all pixels integers
+ sid = np.argmin(test) # fine tune scale
+
+ if scale < 16000 or scale > 16400:
+ warnings.warn("scale not in expected range")
+ print(
+ "guessing scale",
+ scale,
+ test[margin],
+ "fine tuning scale",
+ scale - margin + sid,
+ "dif",
+ test[sid],
+ "time",
+ time.time() - start,
+ )
+
+ scale = 16384
+ else:
+ scale = scale - margin + sid
+ # all the recovered scale values seems to be up to 16384,
+ # we can stretch to 65535 (for better visualization, most tiff viewers expect that range)
+ scale = min(65535, scale * 4)
+ img[i] = im * scale
+
+ img = img.astype(np.uint16)
+ return img
+
+
+def concatenate_masks(mask_dir):
+ labeled_mask = None
+ i = 0
+ for filename in sorted(os.listdir(mask_dir)):
+ if filename.endswith(".png"):
+ mask = imageio.imread(os.path.join(mask_dir, filename))
+ if labeled_mask is None:
+ labeled_mask = np.zeros(shape=mask.shape, dtype=np.uint16)
+ labeled_mask[mask > 0] = i
+ i = i + 1
+
+ if i <= 255:
+ labeled_mask = labeled_mask.astype(np.uint8)
+
+ return labeled_mask
+
+
+def get_filenames_exclude_masks(dir1, target_string):
+ filenames = []
+ # Combine lists of files from both directories
+ files = os.listdir(dir1)
+ # Filter files that contain the target string but exclude 'masks'
+ filenames = [f for f in files if target_string in f and "masks" not in f]
+
+ return filenames
+
+
+def remove_overlaps(masks, medians, overlap_threshold=0.75):
+ """replace overlapping mask pixels with mask id of closest mask
+ if mask fully within another mask, remove it
+ masks = Nmasks x Ly x Lx
+ """
+ cellpix = masks.sum(axis=0)
+ igood = np.ones(masks.shape[0], "bool")
+ for i in masks.sum(axis=(1, 2)).argsort():
+ npix = float(masks[i].sum())
+ noverlap = float(masks[i][cellpix > 1].sum())
+ if noverlap / npix >= overlap_threshold:
+ igood[i] = False
+ cellpix[masks[i] > 0] -= 1
+ # print(cellpix.min())
+ print(f"removing {(~igood).sum()} masks")
+ masks = masks[igood]
+ medians = medians[igood]
+ cellpix = masks.sum(axis=0)
+ overlaps = np.array(np.nonzero(cellpix > 1.0)).T
+ dists = ((overlaps[:, :, np.newaxis] - medians.T) ** 2).sum(axis=1)
+ tocell = np.argmin(dists, axis=1)
+ masks[:, overlaps[:, 0], overlaps[:, 1]] = 0
+ masks[tocell, overlaps[:, 0], overlaps[:, 1]] = 1
+
+ # labels should be 1 to mask.shape[0]
+ masks = masks.astype(int) * np.arange(1, masks.shape[0] + 1, 1, int)[:, np.newaxis, np.newaxis]
+ masks = masks.sum(axis=0)
+ gc.collect()
+ return masks
+
+
+def livecell_process_files(dataset_dir):
+ """
+ This function takes in the directory of livecell extracted dataset as input and
+ extracts labels from the coco format.
+ """
+
+ # "A172", "BT474", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"
+ # "BV2" is being skipped, runs into memory constraints
+ cell_type_list = ["A172", "BT474", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"]
+ for each_cell_tp in cell_type_list:
+ for split in ["train", "val", "test"]:
+ print(f"Working on split: {split}")
+
+ if split == "test":
+ img_path = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp)
+ msk_path = os.path.join(dataset_dir, "images", "livecell_test_images", each_cell_tp + "_masks")
+ else:
+ img_path = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp)
+ msk_path = os.path.join(dataset_dir, "images", "livecell_train_val_images", each_cell_tp + "_masks")
+ if not os.path.exists(msk_path):
+ os.makedirs(msk_path)
+
+ # annotation path
+ path = os.path.join(
+ dataset_dir,
+ "livecell-dataset.s3.eu-central-1.amazonaws.com",
+ "LIVECell_dataset_2021",
+ "annotations",
+ "LIVECell_single_cells",
+ each_cell_tp.lower(),
+ split + ".json",
+ )
+ annotation = COCO(path)
+ # Convert COCO format segmentation to binary mask
+ images = annotation.loadImgs(annotation.getImgIds())
+ height = []
+ width = []
+ for index, im in enumerate(images):
+ print("Status: {}/{}, Process image: {}".format(index, len(images), im["file_name"]))
+ if (
+ im["file_name"] == "BV2_Phase_C4_2_03d00h00m_1.tif"
+ or im["file_name"] == "BV2_Phase_C4_2_03d00h00m_3.tif"
+ ):
+ print("Skipping the file: BV2_Phase_C4_2_03d00h00m_1.tif, as it is troublesome")
+ continue
+ # load image
+ img = Image.open(os.path.join(img_path, im["file_name"])).convert("L")
+ height.append(img.size[0])
+ width.append(img.size[1])
+
+ # load and display instance annotations
+ annids = annotation.getAnnIds(imgIds=im["id"], iscrowd=None)
+ anns = annotation.loadAnns(annids)
+
+ medians = []
+ masks = []
+ k = 0
+ for ann in anns:
+ # convert segmentation to binary mask
+ mask = annotation.annToMask(ann)
+ masks.append(mask)
+ ypix, xpix = mask.nonzero()
+ medians.append(np.array([ypix.mean().astype(np.float32), xpix.mean().astype(np.float32)]))
+ k += 1
+
+ masks = np.array(masks).astype(np.int8)
+ medians = np.array(medians)
+ masks = remove_overlaps(masks, medians, overlap_threshold=0.75)
+ gc.collect()
+
+ # ## Create new name for the image and also for the mask and save them as .tif format
+ # masks_int32 = masks.astype(np.int32)
+ # mask_pil = Image.fromarray(masks_int32, 'I')
+
+ t_filename = im["file_name"]
+ # cell_type = t_filename.split('_')[0] #? not used
+ new_mask_name = t_filename[:-4] + "_masks.tif"
+ # mask_pil.save(os.path.join(msk_path, new_mask_name))
+ imageio.imwrite(os.path.join(msk_path, new_mask_name), min_label_precision(masks))
+ gc.collect()
+
+ print(f"In total {len(images)} images")
+
+
+def tissuenet_process_files(dataset_dir):
+ """
+ This function takes in the directory of TissueNet extracted dataset as input and
+ creates tiled images into 4 from each image
+ """
+
+ for folder in ["train", "val", "test"]:
+ if not os.path.exists(os.path.join(dataset_dir, "tissuenet_1.0", folder)):
+ os.mkdir(os.path.join(dataset_dir, "tissuenet_1.0", folder))
+
+ for folder in ["train", "val", "test"]:
+ print(f"Working on {folder} directory of tissuenet")
+ f_name = f"tissuenet_1.0/tissuenet_v1.0_{folder}.npz"
+ dat = np.load(os.path.join(dataset_dir, f_name))
+ data = dat["X"]
+ labels = dat["y"]
+ tissues = dat["tissue_list"]
+ platforms = dat["platform_list"]
+ tlabels = np.unique(tissues)
+ plabels = np.unique(platforms)
+ tp = 0
+ for t in tlabels:
+ for p in plabels:
+ ix = ((tissues == t) * (platforms == p)).nonzero()[0]
+ tp += 1
+ if len(ix) > 0:
+ print(f"Working on {t} {p}")
+
+ for k, i in enumerate(ix):
+ print(f"Status: {k}/{len(ix)} {tp}/{len(tlabels) * len(plabels)} {t} {p}")
+ img = data[i].transpose(2, 0, 1)
+ label = labels[i][:, :, 0]
+
+ img = guess_convert_to_uint16(img) # guess inverse scale and convert to uint16
+ label = min_label_precision(label)
+
+ if folder == "train":
+ img = img.reshape(2, 2, 256, 2, 256).transpose(0, 1, 3, 2, 4).reshape(2, 4, 256, 256)
+ label = label.reshape(2, 256, 2, 256).transpose(0, 2, 1, 3).reshape(4, 256, 256)
+
+ zero_channel = np.zeros((1, img.shape[1], img.shape[2], img.shape[3]), dtype=img.dtype)
+
+ # Concatenate the zero channel with the original array along the first dimension
+ new_array = np.concatenate([img, zero_channel], axis=0)
+ # reshaped_array = np.transpose(new_array, (1, 2, 3, 0))
+ for j in range(4):
+ img_name = f"{folder}/{t}_{p}_{k}_{j}.tif"
+ mask_name = f"{folder}/{t}_{p}_{k}_{j}_masks.tif"
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", img_name), new_array[:, j])
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", mask_name), label[j])
+ else:
+ zero_channel = np.zeros((1, img.shape[1], img.shape[2]), dtype=img.dtype)
+ new_array = np.concatenate([img, zero_channel], axis=0)
+ # reshaped_array = np.transpose(new_array, (1, 2, 0))
+ img_name = f"{folder}/{t}_{p}_{k}.tif"
+ mask_name = f"{folder}/{t}_{p}_{k}_masks.tif"
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", img_name), new_array)
+ imageio.imwrite(os.path.join(dataset_dir, "tissuenet_1.0", mask_name), label)
+
+
+def kaggle_process_files(dataset_dir):
+ """
+ This function takes in the directory of kaggle nuclei extracted dataset as input and
+ creates a json list with 5 folds.
+ Please note that there are some hard-coded directory names as per the original dataset.
+ The function creates an instance processed dataset and then a 5 fold json file based on
+ the instance processed dataset
+ """
+ data_dir = os.path.join(dataset_dir, "stage1_train")
+ saving_path = os.path.join(dataset_dir, "instance_processed_data")
+ if not os.path.exists(saving_path):
+ os.mkdir(saving_path)
+
+ # Process the images and create instance masks first
+ for idx, subdir in enumerate(os.listdir(data_dir)):
+ subdir_path = os.path.join(data_dir, subdir)
+ if os.path.isdir(subdir_path):
+ images_dir = os.path.join(subdir_path, "images")
+ masks_dir = os.path.join(subdir_path, "masks")
+ if os.path.isdir(images_dir) and os.path.isdir(masks_dir):
+ image_file = os.path.join(images_dir, os.listdir(images_dir)[0])
+ filename_prefix = f"kg_bowl_{idx}_"
+
+ mask_data = concatenate_masks(masks_dir)
+
+ # ## Apply channel-wise normalization and use only the first three channels
+ # image_data = imageio.imread(image_file)
+ # normalized_image = normalize_image(image_data[..., :3])
+ # imageio.imwrite(os.path.join(saving_path, f"{filename_prefix}img.tiff"), normalized_image)
+ shutil.copyfile(image_file, os.path.join(saving_path, f"{filename_prefix}img.png"))
+ imageio.imwrite(os.path.join(saving_path, f"{filename_prefix}img_masks.tiff"), mask_data)
+
+
+def extract_zip(zip_path, extract_to):
+ # Ensure the target directory exists
+ print(f"Extracting from: {zip_path}")
+ print(f"Extracting to: {extract_to}")
+
+ if not os.path.exists(extract_to):
+ os.makedirs(extract_to)
+
+ # Extract all contents of the zip file to the specified directory
+ with zipfile.ZipFile(zip_path, "r") as zip_ref:
+ zip_ref.extractall(extract_to)
+
+
+def main():
+ parser = argparse.ArgumentParser(description="Script to process the cell imaging datasets")
+ parser.add_argument("--dir", type=str, help="Directory of datasets to process it ...", default="/set/the/path")
+
+ args = parser.parse_args()
+ data_root_path = os.path.normpath(args.dir)
+
+ dataset_dict = {
+ "cellpose_dataset": ["train.zip", "test.zip"],
+ "deepbacs_dataset": ["deepbacs.zip"],
+ "kaggle_dataset": ["data-science-bowl-2018.zip"],
+ "nips_dataset": ["nips_train.zip", "nips_test.zip"],
+ "omnipose_dataset": ["datasets.zip"],
+ "tissuenet_dataset": ["tissuenet_v1.0.zip"],
+ "livecell_dataset": [
+ "livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/images_per_celltype.zip"
+ ],
+ }
+
+ for key, value in dataset_dict.items():
+ dataset_path = os.path.join(data_root_path, key)
+
+ for each_zipped in value:
+ in_path = os.path.join(dataset_path, each_zipped)
+ try:
+ if os.path.exists(in_path):
+ print(f"File exists at: {in_path}")
+ except Exception:
+ print(f"File: {in_path} was not found")
+ out_path = os.path.join(dataset_path)
+ extract_zip(in_path, out_path)
+
+ print("If we reached here, that means all zip files got extracted ... Working on pre-processing")
+
+ # Looping over all datasets again, Cellpose & Deepbacs have a similar directory structure
+ for key, _value in dataset_dict.items():
+ if key == "kaggle_dataset":
+ print("Needs additional extraction")
+ train_zip_path = os.path.join(data_root_path, key, "stage1_train.zip")
+ zip_out_path = os.path.join(data_root_path, key, "stage1_train")
+ extract_zip(train_zip_path, zip_out_path)
+ print("Processing Kaggle Dataset ...")
+ dataset_path = os.path.join(data_root_path, key)
+ kaggle_process_files(dataset_dir=dataset_path)
+
+ elif key == "livecell_dataset":
+ print("Processing LiveCell Dataset ...")
+ print(
+ "Fyi, this processing might take upto an hour, coffee break might be more fruitful in the meanwhile ..."
+ )
+ dataset_path = os.path.join(data_root_path, key)
+ livecell_process_files(dataset_dir=dataset_path)
+
+ elif key == "tissuenet_dataset":
+ print("Processing TissueNet Dataset ...")
+ dataset_path = os.path.join(data_root_path, key)
+ tissuenet_process_files(dataset_dir=dataset_path)
+
+ return None
+
+
+if __name__ == "__main__":
+ main()
diff --git a/models/vista2d/download_preprocessor/readme.md b/models/vista2d/download_preprocessor/readme.md
new file mode 100644
index 00000000..e07cc8be
--- /dev/null
+++ b/models/vista2d/download_preprocessor/readme.md
@@ -0,0 +1,73 @@
+## Tutorial: VISTA2D Model Creation
+
+This tutorial will guide the users to setting up all the datasets, running pre-processing, creation of organized json file lists which can be provided to VISTA-2D training pipeline.
+Some datasets need to be manually downloaded, others will be downloaded by a provided script. Please do not manually unzip any of the downloaded files, it will be automatically handled in the final step.
+
+### List of Datasets
+1.) [Cellpose](https://www.cellpose.org/dataset)
+
+2.) [TissueNet](https://datasets.deepcell.org/login)
+
+3.) [Kaggle Nuclei Segmentation](https://www.kaggle.com/c/data-science-bowl-2018/data)
+
+4.) [Omnipose - OSF repository](https://osf.io/xmury/)
+
+5.) [NIPS Cell Segmentation Challenge](https://neurips22-cellseg.grand-challenge.org/)
+
+6.) [LiveCell](https://sartorius-research.github.io/LIVECell/)
+
+7.) [Deepbacs](https://github.com/HenriquesLab/DeepBacs/wiki/Segmentation)
+
+Datasets 1-4 need to be manually downloaded, instructions to download them have been provided below.
+
+### Manual Dataset Download Instructions
+#### 1.) Cellpose:
+The dataset can be downloaded from this [link](https://www.cellpose.org/dataset). Please see below screenshots to assist in downloading it
+
+Please enter your email and accept terms and conditions to download the dataset.
+
+
+Click on train.zip and test.zip to download both directories independently. They both need to be placed in a `cellpose_dataset` directory. The `cellpose_dataset` will have to be created by the user in the root data directory.
+
+#### 2.) TissueNet
+Login credentials have to be created at below provided link. Please see below screenshots for further assistance.
+
+
+Please create an account at the provided [link](https://datasets.deepcell.org/login).
+
+
+After logging in, the above page will be visible, please make sure that version 1.0 is selected for TissueNet before clicking on download button.
+All the downloaded files need to be placed in a `tissuenet_dataset` directory, this directory has to be created by the user.
+
+#### 3.) Kaggle Nuclei Segmentation
+Kaggle credentials are required in order to access this dataset at this [link](https://www.kaggle.com/c/data-science-bowl-2018/data), the user will have to register for the challenge to access and download the dataset.
+Please refer below screenshots for additional help.
+
+
+The `Download All` button needs to be used so all files are downloaded, the files need to be placed in a directory created by the user `kaggle_dataset`.
+
+#### 4.) Omnipose
+The Omnipose dataset is hosted on an [OSF repository](https://osf.io/xmury/) and the dataset part needs to be downloaded from it. Please refer below screenshots for further assistance.
+
+
+The `datasets` directory needs to be selected as highlighted in the screenshot, then `download as zip` needs to be pressed for downloading the dataset. The user will have to place all the files in
+a user created directory named `omnipose_dataset`.
+
+### The remaining datasets will be downloaded by a python script.
+To run the script use the following example command `python all_file_downloader.py --download_path provide_the_same_root_data_path`
+
+After completion of downloading of all datasets, below is how the data root directory should look:
+
+
+
+### Process the downloaded data
+To execute VISTA-2D training pipeline, some datasets require label conversion. Please use the `root_data_path` as the input to the script, example command to execute the script is given below:
+
+`python generate_json.py --data_root provide_the_same_root_data_path`
+
+### Generation of Json data lists (Optional)
+If one desires to generate JSON files from scratch, `generate_json.py` script performs both processing and creation of JSON files.
+To execute VISTA-2D training pipeline, some datasets require label conversion and then a json file list which the VISTA-2D training uses a format.
+Creating the json lists from the raw dataset sources, please use the `root_data_path` as the input to the script, example command to execute the script is given below:
+
+`python generate_json.py --data_root provide_the_same_root_data_path`
diff --git a/models/vista2d/download_preprocessor/tissuenet_download.png b/models/vista2d/download_preprocessor/tissuenet_download.png
new file mode 100644
index 00000000..f078fe9b
Binary files /dev/null and b/models/vista2d/download_preprocessor/tissuenet_download.png differ
diff --git a/models/vista2d/download_preprocessor/tissuenet_login.png b/models/vista2d/download_preprocessor/tissuenet_login.png
new file mode 100644
index 00000000..1fcf1f23
Binary files /dev/null and b/models/vista2d/download_preprocessor/tissuenet_login.png differ
diff --git a/models/vista2d/download_preprocessor/urls.txt b/models/vista2d/download_preprocessor/urls.txt
new file mode 100644
index 00000000..b85707f2
--- /dev/null
+++ b/models/vista2d/download_preprocessor/urls.txt
@@ -0,0 +1,39 @@
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LICENSE
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/a172/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bt474/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/bv2/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/huh7/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/mcf7/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/shsy5y/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skbr3/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/test.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/train.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/annotations/LIVECell_single_cells/skov3/val.json
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/LIVECell_dataset_2021/images_per_celltype.zip
+http://livecell-dataset.s3.eu-central-1.amazonaws.com/README.md
diff --git a/models/vista2d/large_files.yml b/models/vista2d/large_files.yml
new file mode 100644
index 00000000..75fe41d5
--- /dev/null
+++ b/models/vista2d/large_files.yml
@@ -0,0 +1,7 @@
+large_files:
+- path: "models/sam_vit_b_01ec64.pth"
+ url: "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth"
+- path: "models/model.pt"
+ url: "https://drive.google.com/file/d/1odLhoOtlxxbEyRq-gvenP8bC0-mw63ng/view?usp=drive_link"
+- path: "datalists.zip"
+ url: "https://github.com/Project-MONAI/model-zoo/releases/download/model_zoo_bundle_data/vista2d_datalists.zip"
diff --git a/models/vista2d/scripts/__init__.py b/models/vista2d/scripts/__init__.py
new file mode 100644
index 00000000..1e97f894
--- /dev/null
+++ b/models/vista2d/scripts/__init__.py
@@ -0,0 +1,10 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
diff --git a/models/vista2d/scripts/cell_distributed_weighted_sampler.py b/models/vista2d/scripts/cell_distributed_weighted_sampler.py
new file mode 100644
index 00000000..6e690b9d
--- /dev/null
+++ b/models/vista2d/scripts/cell_distributed_weighted_sampler.py
@@ -0,0 +1,119 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+# based on Pytorch DistributedSampler and WeightedRandomSampler combined
+
+import math
+from typing import Iterator, Optional, Sequence, TypeVar
+
+import torch
+import torch.distributed as dist
+from torch.utils.data import Dataset, Sampler
+
+__all__ = ["DistributedWeightedSampler"]
+
+T_co = TypeVar("T_co", covariant=True)
+
+
+class DistributedWeightedSampler(Sampler[T_co]):
+ def __init__(
+ self,
+ dataset: Dataset,
+ weights: Sequence[float],
+ num_samples: int,
+ num_replicas: Optional[int] = None,
+ rank: Optional[int] = None,
+ shuffle: bool = True,
+ seed: int = 0,
+ drop_last: bool = False,
+ ) -> None:
+ if not isinstance(num_samples, int) or isinstance(num_samples, bool) or num_samples <= 0:
+ raise ValueError(f"num_samples should be a positive integer value, but got num_samples={num_samples}")
+
+ weights_tensor = torch.as_tensor(weights, dtype=torch.float)
+ if len(weights_tensor.shape) != 1:
+ raise ValueError(
+ "weights should be a 1d sequence but given " f"weights have shape {tuple(weights_tensor.shape)}"
+ )
+
+ self.weights = weights_tensor
+ self.num_samples = num_samples
+
+ if num_replicas is None:
+ if not dist.is_available():
+ raise RuntimeError("Requires distributed package to be available")
+ num_replicas = dist.get_world_size()
+ if rank is None:
+ if not dist.is_available():
+ raise RuntimeError("Requires distributed package to be available")
+ rank = dist.get_rank()
+ if rank >= num_replicas or rank < 0:
+ raise ValueError(f"Invalid rank {rank}, rank should be in the interval [0, {num_replicas - 1}]")
+ self.dataset = dataset
+ self.num_replicas = num_replicas
+ self.rank = rank
+ self.epoch = 0
+ self.drop_last = drop_last
+ self.shuffle = shuffle
+
+ if self.shuffle:
+ self.num_samples = int(math.ceil(self.num_samples / self.num_replicas))
+ else:
+ # this is not used, as we always shuffle, the only reason to use this class
+
+ # If the dataset length is evenly divisible by # of replicas, then there
+ # is no need to drop any data, since the dataset will be split equally.
+ if self.drop_last and len(self.dataset) % self.num_replicas != 0: # type: ignore[arg-type]
+ # Split to nearest available length that is evenly divisible.
+ # This is to ensure each rank receives the same amount of data when
+ # using this Sampler.
+ self.num_samples = math.ceil(
+ (len(self.dataset) - self.num_replicas) / self.num_replicas # type: ignore[arg-type]
+ )
+ else:
+ self.num_samples = math.ceil(len(self.dataset) / self.num_replicas) # type: ignore[arg-type]
+
+ self.total_size = self.num_samples * self.num_replicas
+ self.shuffle = shuffle
+ self.seed = seed
+
+ def __iter__(self) -> Iterator[T_co]:
+ if self.shuffle:
+ # deterministically shuffle based on epoch and seed
+ g = torch.Generator()
+ g.manual_seed(self.seed + self.epoch)
+ indices = torch.multinomial(input=self.weights, num_samples=self.total_size, replacement=True, generator=g).tolist() # type: ignore[arg-type]
+ else:
+ # this is not used, as we always shuffle, the only reason to use this class
+ indices = list(range(len(self.dataset))) # type: ignore[arg-type]
+ if not self.drop_last:
+ # add extra samples to make it evenly divisible
+ padding_size = self.total_size - len(indices)
+ if padding_size <= len(indices):
+ indices += indices[:padding_size]
+ else:
+ indices += (indices * math.ceil(padding_size / len(indices)))[:padding_size]
+ else:
+ # remove tail of data to make it evenly divisible.
+ indices = indices[: self.total_size]
+ assert len(indices) == self.total_size
+
+ # subsample
+ indices = indices[self.rank : self.total_size : self.num_replicas]
+ assert len(indices) == self.num_samples
+
+ return iter(indices)
+
+ def __len__(self) -> int:
+ return self.num_samples
+
+ def set_epoch(self, epoch: int) -> None:
+ self.epoch = epoch
diff --git a/models/vista2d/scripts/cell_sam_wrapper.py b/models/vista2d/scripts/cell_sam_wrapper.py
new file mode 100644
index 00000000..9c13676d
--- /dev/null
+++ b/models/vista2d/scripts/cell_sam_wrapper.py
@@ -0,0 +1,72 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import torch
+from segment_anything.build_sam import build_sam_vit_b
+from torch import nn
+from torch.nn import functional as F
+
+
+class CellSamWrapper(torch.nn.Module):
+ def __init__(
+ self,
+ auto_resize_inputs=True,
+ network_resize_roi=(1024, 1024),
+ checkpoint="sam_vit_b_01ec64.pth",
+ return_features=False,
+ *args,
+ **kwargs,
+ ) -> None:
+ super().__init__(*args, **kwargs)
+
+ print(
+ f"CellSamWrapper auto_resize_inputs {auto_resize_inputs} "
+ f"network_resize_roi {network_resize_roi} "
+ f"checkpoint {checkpoint}"
+ )
+ self.network_resize_roi = network_resize_roi
+ self.auto_resize_inputs = auto_resize_inputs
+ self.return_features = return_features
+
+ model = build_sam_vit_b(checkpoint=checkpoint)
+
+ model.prompt_encoder = None
+ model.mask_decoder = None
+
+ model.mask_decoder = nn.Sequential(
+ nn.BatchNorm2d(num_features=256),
+ nn.ReLU(inplace=True),
+ nn.ConvTranspose2d(256, 128, kernel_size=3, stride=2, padding=1, output_padding=1, bias=False),
+ nn.BatchNorm2d(num_features=128),
+ nn.ReLU(inplace=True),
+ nn.ConvTranspose2d(128, 3, kernel_size=3, stride=2, padding=1, output_padding=1, bias=True),
+ )
+
+ self.model = model
+
+ def forward(self, x):
+ # print("CellSamWrapper x0", x.shape)
+ sh = x.shape[2:]
+
+ if self.auto_resize_inputs:
+ x = F.interpolate(x, size=self.network_resize_roi, mode="bilinear")
+
+ # print("CellSamWrapper x1", x.shape)
+ x = self.model.image_encoder(x) # shape: (1, 256, 64, 64)
+ # print("CellSamWrapper image_embeddings", x.shape)
+
+ if not self.return_features:
+ x = self.model.mask_decoder(x)
+ if self.auto_resize_inputs:
+ x = F.interpolate(x, size=sh, mode="bilinear")
+
+ # print("CellSamWrapper x final", x.shape)
+ return x
diff --git a/models/vista2d/scripts/components.py b/models/vista2d/scripts/components.py
new file mode 100644
index 00000000..454356c0
--- /dev/null
+++ b/models/vista2d/scripts/components.py
@@ -0,0 +1,298 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import json
+import os
+
+import cv2
+import fastremap
+import numpy as np
+import PIL
+import tifffile
+import torch
+import torch.nn.functional as F
+from cellpose.dynamics import compute_masks, masks_to_flows
+from cellpose.metrics import _intersection_over_union, _true_positive
+from monai.apps import get_logger
+from monai.data import MetaTensor
+from monai.transforms import MapTransform
+from monai.utils import ImageMetaKey, convert_to_dst_type
+
+logger = get_logger("VistaCell")
+
+
+class LoadTiffd(MapTransform):
+ def __call__(self, data):
+ d = dict(data)
+ for key in self.key_iterator(d):
+ filename = d[key]
+
+ extension = os.path.splitext(filename)[1][1:]
+ image_size = None
+
+ if extension in ["tif", "tiff"]:
+ img_array = tifffile.imread(filename) # use tifffile for tif images
+ image_size = img_array.shape
+ if len(img_array.shape) == 3 and img_array.shape[-1] <= 3:
+ img_array = np.transpose(img_array, (2, 0, 1)) # channels first without transpose
+ else:
+ img_array = np.array(PIL.Image.open(filename)) # PIL for all other images (png, jpeg)
+ image_size = img_array.shape
+ if len(img_array.shape) == 3:
+ img_array = np.transpose(img_array, (2, 0, 1)) # channels first
+
+ if len(img_array.shape) not in [2, 3]:
+ raise ValueError(
+ "Unsupported image dimensions, filename " + str(filename) + " shape " + str(img_array.shape)
+ )
+
+ if len(img_array.shape) == 2:
+ img_array = img_array[np.newaxis] # add channels_first if no channel
+
+ if key == "label":
+ if img_array.shape[0] > 1:
+ print(
+ f"Strange case, label with several channels {filename} shape {img_array.shape}, keeping only first"
+ )
+ img_array = img_array[[0]]
+
+ elif key == "image":
+ if img_array.shape[0] == 1:
+ img_array = np.repeat(img_array, repeats=3, axis=0) # if grayscale, repeat as 3 channels
+ elif img_array.shape[0] == 2:
+ print(
+ f"Strange case, image with 2 channels {filename} shape {img_array.shape}, appending first channel to make 3"
+ )
+ img_array = np.stack(
+ (img_array[0], img_array[1], img_array[0]), axis=0
+ ) # this should not happen, we got 2 channel input image
+ elif img_array.shape[0] > 3:
+ print(f"Strange case, image with >3 channels, {filename} shape {img_array.shape}, keeping first 3")
+ img_array = img_array[:3]
+
+ meta_data = {ImageMetaKey.FILENAME_OR_OBJ: filename, ImageMetaKey.SPATIAL_SHAPE: image_size}
+ d[key] = MetaTensor.ensure_torch_and_prune_meta(img_array, meta_data)
+
+ return d
+
+
+class SaveTiffd(MapTransform):
+ def __init__(self, output_dir, data_root_dir="/", nested_folder=False, *args, **kwargs) -> None:
+ super().__init__(*args, **kwargs)
+
+ self.output_dir = output_dir
+ self.data_root_dir = data_root_dir
+ self.nested_folder = nested_folder
+
+ def set_data_root_dir(self, data_root_dir):
+ self.data_root_dir = data_root_dir
+
+ def __call__(self, data):
+ d = dict(data)
+ os.makedirs(self.output_dir, exist_ok=True)
+
+ for key in self.key_iterator(d):
+ seg = d[key]
+ filename = seg.meta[ImageMetaKey.FILENAME_OR_OBJ]
+
+ basename = os.path.splitext(os.path.basename(filename))[0]
+
+ if self.nested_folder:
+ reldir = os.path.relpath(os.path.dirname(filename), self.data_root_dir)
+ outdir = os.path.join(self.output_dir, reldir)
+ os.makedirs(outdir, exist_ok=True)
+ else:
+ outdir = self.output_dir
+
+ outname = os.path.join(outdir, basename + ".tif")
+
+ label = seg.cpu().numpy()
+ lm = label.max()
+ if lm <= 255:
+ label = label.astype(np.uint8)
+ elif lm <= 65535:
+ label = label.astype(np.uint16)
+ else:
+ label = label.astype(np.uint32)
+
+ tifffile.imwrite(outname, label)
+
+ print(f"Saving {outname} shape {label.shape} max {label.max()} dtype {label.dtype}")
+
+ return d
+
+
+class LabelsToFlows(MapTransform):
+ # based on dynamics labels_to_flows()
+ # created a 3 channel output (foreground, flowx, flowy) and saves under flow (new) key
+
+ def __init__(self, flow_key, *args, **kwargs) -> None:
+ super().__init__(*args, **kwargs)
+ self.flow_key = flow_key
+
+ def __call__(self, data):
+ d = dict(data)
+ for key in self.key_iterator(d):
+ label = d[key].int().numpy()
+
+ label = fastremap.renumber(label, in_place=True)[0]
+ veci = masks_to_flows(label[0], device=None)
+
+ flows = np.concatenate((label > 0.5, veci), axis=0).astype(np.float32)
+ flows = convert_to_dst_type(flows, d[key], dtype=torch.float, device=d[key].device)[0]
+ d[self.flow_key] = flows
+ # meta_data = {ImageMetaKey.FILENAME_OR_OBJ : filename}
+ # d[key] = MetaTensor.ensure_torch_and_prune_meta(img_array, meta_data)
+ return d
+
+
+class LogitsToLabels:
+ def __call__(self, logits, filename=None):
+ device = logits.device
+ logits = logits.float().cpu().numpy()
+ dp = logits[1:] # vectors
+ cellprob = logits[0] # foreground prob (logit)
+
+ try:
+ pred_mask, p = compute_masks(
+ dp, cellprob, niter=200, cellprob_threshold=0.4, flow_threshold=0.4, interp=True, device=device
+ )
+ except RuntimeError as e:
+ logger.warning(f"compute_masks failed on GPU retrying on CPU {logits.shape} file {filename} {e}")
+ pred_mask, p = compute_masks(
+ dp, cellprob, niter=200, cellprob_threshold=0.4, flow_threshold=0.4, interp=True, device=None
+ )
+
+ return pred_mask, p
+
+
+class LogitsToLabelsd(MapTransform):
+ def __call__(self, data):
+ d = dict(data)
+ f = LogitsToLabels()
+ for key in self.key_iterator(d):
+ pred_mask, p = f(d[key])
+ d[key] = pred_mask
+ d[f"{key}_centroids"] = p
+ return d
+
+
+class SaveTiffExd(MapTransform):
+ def __init__(self, output_dir, output_ext=".png", output_postfix="seg", image_key="image", *args, **kwargs) -> None:
+ super().__init__(*args, **kwargs)
+
+ self.output_dir = output_dir
+ self.output_ext = output_ext
+ self.output_postfix = output_postfix
+ self.image_key = image_key
+
+ def to_polygons(self, contours):
+ polygons = []
+ for contour in contours:
+ if len(contour) < 3:
+ continue
+ polygons.append(np.squeeze(contour).astype(int).tolist())
+ return polygons
+
+ def __call__(self, data):
+ d = dict(data)
+
+ output_dir = d.get("output_dir", self.output_dir)
+ output_ext = d.get("output_ext", self.output_ext)
+ overlayed_masks = d.get("overlayed_masks", False)
+ output_contours = d.get("output_contours", False)
+
+ os.makedirs(self.output_dir, exist_ok=True)
+
+ img = d.get(self.image_key, None)
+ filename = img.meta.get(ImageMetaKey.FILENAME_OR_OBJ) if img is not None else None
+ image_size = img.meta.get(ImageMetaKey.SPATIAL_SHAPE) if img is not None else None
+ basename = os.path.splitext(os.path.basename(filename))[0] if filename else "mask"
+ logger.info(f"File: {filename}; Base: {basename}")
+
+ for key in self.key_iterator(d):
+ label = d[key]
+ output_filename = f"{basename}{'_' + self.output_postfix if self.output_postfix else ''}{output_ext}"
+ output_filepath = os.path.join(output_dir, output_filename)
+ lm = label.max()
+ logger.info(f"Mask Shape: {label.shape}; Instances: {lm}")
+
+ if lm <= 255:
+ label = label.astype(np.uint8)
+ elif lm <= 65535:
+ label = label.astype(np.uint16)
+ else:
+ label = label.astype(np.uint32)
+
+ tifffile.imwrite(output_filepath, label)
+ logger.info(f"Saving {output_filepath}")
+
+ polygons = []
+ if overlayed_masks:
+ logger.info(f"Overlay Masks: Reading original Image: {filename}")
+ image = cv2.imread(filename)
+ mask = cv2.imread(output_filepath, 0)
+
+ for i in range(1, np.max(mask)):
+ m = np.zeros_like(mask)
+ m[mask == i] = 1
+ color = np.random.choice(range(256), size=3).tolist()
+ contours, _ = cv2.findContours(m, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
+ polygons.extend(self.to_polygons(contours))
+ cv2.drawContours(image, contours, -1, color, 1)
+ cv2.imwrite(output_filepath, image)
+ logger.info(f"Overlay Masks: Saving {output_filepath}")
+ else:
+ contours, _ = cv2.findContours(label, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
+ polygons.extend(self.to_polygons(contours))
+
+ meta_json = {"image_size": image_size, "contours": len(polygons)}
+ with open(os.path.join(output_dir, "meta.json"), "w") as fp:
+ json.dump(meta_json, fp, indent=2)
+
+ if output_contours:
+ logger.info(f"Total Polygons: {len(polygons)}")
+ with open(os.path.join(output_dir, "contours.json"), "w") as fp:
+ json.dump({"count": len(polygons), "contours": polygons}, fp, indent=2)
+
+ return d
+
+
+# Loss (adopted from Cellpose)
+class CellLoss:
+ def __call__(self, y_pred, y):
+ loss = 0.5 * F.mse_loss(y_pred[:, 1:], 5 * y[:, 1:]) + F.binary_cross_entropy_with_logits(
+ y_pred[:, [0]], y[:, [0]]
+ )
+ return loss
+
+
+# Accuracy (adopted from Cellpose)
+class CellAcc:
+ def __call__(self, mask_pred, mask_true):
+ if isinstance(mask_true, torch.Tensor):
+ mask_true = mask_true.cpu().numpy()
+
+ if isinstance(mask_pred, torch.Tensor):
+ mask_pred = mask_pred.cpu().numpy()
+
+ # print("CellAcc mask_true", mask_true.shape, 'max', np.max(mask_true), ",
+ # "'mask_pred', mask_pred.shape, 'max', np.max(mask_pred) )
+
+ iou = _intersection_over_union(mask_true, mask_pred)[1:, 1:]
+ tp = _true_positive(iou, th=0.5)
+
+ fp = np.max(mask_pred) - tp
+ fn = np.max(mask_true) - tp
+ ap = tp / (tp + fp + fn)
+
+ # print("CellAcc ap", ap, 'tp', tp, 'fp', fp, 'fn', fn)
+ return ap
diff --git a/models/vista2d/scripts/utils.py b/models/vista2d/scripts/utils.py
new file mode 100644
index 00000000..ea50e509
--- /dev/null
+++ b/models/vista2d/scripts/utils.py
@@ -0,0 +1,86 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import logging
+import os
+import warnings
+from logging.config import fileConfig
+from pathlib import Path
+
+import numpy as np
+from monai.apps import get_logger
+from monai.apps.utils import DEFAULT_FMT
+from monai.bundle import ConfigParser
+from monai.utils import RankFilter, ensure_tuple
+
+logger = get_logger("VistaCell")
+
+np.set_printoptions(formatter={"float": "{: 0.3f}".format}, suppress=True)
+logging.getLogger("torch.nn.parallel.distributed").setLevel(logging.WARNING)
+warnings.filterwarnings("ignore", message=".*Divide by zero.*") # intensity transform divide by zero warning
+
+LOGGING_CONFIG = {
+ "version": 1,
+ "disable_existing_loggers": False,
+ "formatters": {"monai_default": {"format": DEFAULT_FMT}},
+ "loggers": {"VistaCell": {"handlers": ["file", "console"], "level": "DEBUG", "propagate": False}},
+ "filters": {"rank_filter": {"()": RankFilter}},
+ "handlers": {
+ "file": {
+ "class": "logging.FileHandler",
+ "filename": "default.log",
+ "mode": "a", # append or overwrite
+ "level": "DEBUG",
+ "formatter": "monai_default",
+ "filters": ["rank_filter"],
+ },
+ "console": {
+ "class": "logging.StreamHandler",
+ "level": "INFO",
+ "formatter": "monai_default",
+ "filters": ["rank_filter"],
+ },
+ },
+}
+
+
+def parsing_bundle_config(config_file, logging_file=None, meta_file=None):
+ if config_file is not None:
+ _config_files = ensure_tuple(config_file)
+ config_root_path = Path(_config_files[0]).parent
+ for _config_file in _config_files:
+ _config_file = Path(_config_file)
+ if _config_file.parent != config_root_path:
+ logger.warning(
+ f"Not all config files are in '{config_root_path}'. If logging_file and meta_file are"
+ f"not specified, '{config_root_path}' will be used as the default config root directory."
+ )
+ if not _config_file.is_file():
+ raise FileNotFoundError(f"Cannot find the config file: {_config_file}.")
+ else:
+ config_root_path = Path("configs")
+
+ logging_file = str(config_root_path / "logging.conf") if logging_file is None else logging_file
+ if os.path.exists(logging_file):
+ fileConfig(logging_file, disable_existing_loggers=False)
+
+ parser = ConfigParser()
+ parser.read_config(config_file)
+ meta_file = str(config_root_path / "metadata.json") if meta_file is None else meta_file
+ if isinstance(meta_file, str) and not os.path.exists(meta_file):
+ logger.error(
+ f"Cannot find the metadata config file: {meta_file}. "
+ "Please see: https://docs.monai.io/en/stable/mb_specification.html"
+ )
+ else:
+ parser.read_meta(f=meta_file)
+
+ return parser
diff --git a/models/vista2d/scripts/workflow.py b/models/vista2d/scripts/workflow.py
new file mode 100644
index 00000000..e987a2f6
--- /dev/null
+++ b/models/vista2d/scripts/workflow.py
@@ -0,0 +1,1205 @@
+# Copyright (c) MONAI Consortium
+# Licensed under the Apache License, Version 2.0 (the "License");
+# you may not use this file except in compliance with the License.
+# You may obtain a copy of the License at
+# http://www.apache.org/licenses/LICENSE-2.0
+# Unless required by applicable law or agreed to in writing, software
+# distributed under the License is distributed on an "AS IS" BASIS,
+# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
+# See the License for the specific language governing permissions and
+# limitations under the License.
+
+import csv
+import gc
+import logging
+import os
+import shutil
+import sys
+import time
+from collections import OrderedDict
+from datetime import datetime
+
+import monai.transforms as mt
+import numpy as np
+import torch
+import torch.distributed as dist
+import yaml
+from monai.apps import get_logger
+from monai.auto3dseg.utils import datafold_read
+from monai.bundle import BundleWorkflow, ConfigParser
+from monai.config import print_config
+from monai.data import DataLoader, Dataset, decollate_batch
+from monai.metrics import CumulativeAverage
+from monai.utils import (
+ BundleProperty,
+ ImageMetaKey,
+ convert_to_dst_type,
+ ensure_tuple,
+ look_up_option,
+ optional_import,
+ set_determinism,
+)
+from torch.cuda.amp import GradScaler, autocast
+from torch.utils.data import WeightedRandomSampler
+from torch.utils.data.distributed import DistributedSampler
+from torch.utils.tensorboard import SummaryWriter
+
+mlflow, mlflow_is_imported = optional_import("mlflow")
+
+
+if __package__ in (None, ""):
+ from cell_distributed_weighted_sampler import DistributedWeightedSampler
+ from components import LabelsToFlows, LoadTiffd, LogitsToLabels
+ from utils import LOGGING_CONFIG, parsing_bundle_config # type: ignore
+else:
+ from .cell_distributed_weighted_sampler import DistributedWeightedSampler
+ from .components import LabelsToFlows, LoadTiffd, LogitsToLabels
+ from .utils import LOGGING_CONFIG, parsing_bundle_config
+
+
+logger = get_logger("VistaCell")
+
+
+class VistaCell(BundleWorkflow):
+ """
+ Primary vista model training workflow that extends
+ monai.bundle.BundleWorkflow for cell segmentation.
+ """
+
+ def __init__(self, config_file=None, meta_file=None, logging_file=None, workflow_type="train", **override):
+ """
+ config_file can be one or a list of config files.
+ the rest key-values in the `override` are to override config content.
+ """
+
+ parser = parsing_bundle_config(config_file, logging_file=logging_file, meta_file=meta_file)
+ parser.update(pairs=override)
+
+ mode = parser.get("mode", None)
+ if mode is not None: # if user specified a `mode` it'll override the workflow_type arg
+ workflow_type = mode
+ else:
+ mode = workflow_type # if user didn't specify mode, the workflow_type will be used
+ super().__init__(workflow_type=workflow_type)
+ self._props = {}
+ self._set_props = {}
+ self.parser = parser
+
+ self.rank = int(os.getenv("LOCAL_RANK", "0"))
+ self.global_rank = int(os.getenv("RANK", "0"))
+ self.is_distributed = dist.is_available() and dist.is_initialized()
+
+ # check if torchrun or bcprun started it
+ if dist.is_torchelastic_launched() or (
+ os.getenv("NGC_ARRAY_SIZE") is not None and int(os.getenv("NGC_ARRAY_SIZE")) > 1
+ ):
+ if dist.is_available():
+ dist.init_process_group(backend="nccl", init_method="env://")
+
+ self.is_distributed = dist.is_available() and dist.is_initialized()
+
+ torch.cuda.set_device(self.config("device"))
+ dist.barrier()
+
+ else:
+ self.is_distributed = False
+
+ if self.global_rank == 0 and self.config("ckpt_path") and not os.path.exists(self.config("ckpt_path")):
+ os.makedirs(self.config("ckpt_path"), exist_ok=True)
+
+ if self.rank == 0:
+ # make sure the log file exists, as a workaround for mult-gpu logging race condition
+ _log_file = self.config("log_output_file", "vista_cell.log")
+ _log_file_dir = os.path.dirname(_log_file)
+ if _log_file_dir and not os.path.exists(_log_file_dir):
+ os.makedirs(_log_file_dir, exist_ok=True)
+
+ print_config()
+
+ if self.is_distributed:
+ dist.barrier()
+
+ seed = self.config("seed", None)
+ if seed is not None:
+ set_determinism(seed)
+ logger.info(f"set determinism seed: {self.config('seed', None)}")
+ elif torch.cuda.is_available():
+ torch.backends.cudnn.benchmark = True
+ logger.info("No seed provided, using cudnn.benchmark for performance.")
+
+ if os.path.exists(self.config("ckpt_path")):
+ self.parser.export_config_file(
+ self.parser.config,
+ os.path.join(self.config("ckpt_path"), "working.yaml"),
+ fmt="yaml",
+ default_flow_style=None,
+ )
+
+ self.add_property("network", required=True)
+ self.add_property("train_loader", required=True)
+ self.add_property("val_dataset", required=False)
+ self.add_property("val_loader", required=False)
+ self.add_property("val_preprocessing", required=False)
+ self.add_property("train_sampler", required=True)
+ self.add_property("val_sampler", required=True)
+ self.add_property("mode", required=False)
+ # set evaluator as required when mode is infer or eval
+ # will change after we enhance the bundle properties
+ self.evaluator = None
+
+ def _set_property(self, name, property, value):
+ # stores user-reset initialized objects that should not be re-initialized.
+ self._set_props[name] = value
+
+ def _get_property(self, name, property):
+ """
+ The customized bundle workflow must implement required properties in:
+ https://github.com/Project-MONAI/MONAI/blob/dev/monai/bundle/properties.py.
+ """
+ if name in self._set_props:
+ self._props[name] = self._set_props[name]
+ return self._props[name]
+ if name in self._props:
+ return self._props[name]
+ try:
+ value = getattr(self, f"get_{name}")()
+ except AttributeError as err:
+ if property[BundleProperty.REQUIRED]:
+ raise ValueError(
+ f"Property '{name}' is required by the bundle format, "
+ f"but the method 'get_{name}' is not implemented."
+ ) from err
+ raise AttributeError from err
+ self._props[name] = value
+ return value
+
+ def config(self, name, default="null", **kwargs):
+ """read the parsed content (evaluate the expression) from the config file."""
+ if default != "null":
+ return self.parser.get_parsed_content(name, default=default, **kwargs)
+ return self.parser.get_parsed_content(name, **kwargs)
+
+ def initialize(self):
+ _log_file = self.config("log_output_file", "vista_cell.log")
+ if _log_file is None:
+ LOGGING_CONFIG["loggers"]["VistaCell"]["handlers"].remove("file")
+ LOGGING_CONFIG["handlers"].pop("file", None)
+ else:
+ LOGGING_CONFIG["handlers"]["file"]["filename"] = _log_file
+ logging.config.dictConfig(LOGGING_CONFIG)
+
+ def get_mode(self):
+ mode_str = self.config("mode", self.workflow_type)
+ return look_up_option(mode_str, ("train", "training", "infer", "inference", "eval", "evaluation"))
+
+ def run(self):
+ if str(self.mode).startswith("train"):
+ return self.train()
+ if str(self.mode).startswith("infer"):
+ return self.infer()
+ return self.validate()
+
+ def finalize(self):
+ if self.is_distributed:
+ dist.destroy_process_group()
+ set_determinism(None)
+
+ def get_network_def(self):
+ return self.config("network_def")
+
+ def get_network(self):
+ pretrained_ckpt_name = self.config("pretrained_ckpt_name", None)
+ pretrained_ckpt_path = self.config("pretrained_ckpt_path", None)
+ if pretrained_ckpt_name is not None and pretrained_ckpt_path is None:
+ # if relative name specified, append to default ckpt_path dir
+ pretrained_ckpt_path = os.path.join(self.config("ckpt_path"), pretrained_ckpt_name)
+
+ if pretrained_ckpt_path is not None and not os.path.exists(pretrained_ckpt_path):
+ logger.info(f"Pretrained checkpoint {pretrained_ckpt_path} not found.")
+ raise ValueError(f"Pretrained checkpoint {pretrained_ckpt_path} not found.")
+
+ if pretrained_ckpt_path is not None and os.path.exists(pretrained_ckpt_path):
+ # not loading sam weights, if we're using our own checkpoint
+ if "checkpoint" in self.parser.config["network_def"]:
+ self.parser.config["network_def"]["checkpoint"] = None
+ model = self.config("network")
+ self.checkpoint_load(ckpt=pretrained_ckpt_path, model=model)
+ else:
+ model = self.config("network")
+
+ if self.config("channels_last", False):
+ model = model.to(memory_format=torch.channels_last)
+
+ if self.is_distributed:
+ model = torch.nn.SyncBatchNorm.convert_sync_batchnorm(model)
+
+ if self.config("compile", False):
+ model = torch.compile(model)
+
+ if self.is_distributed:
+ model = torch.nn.parallel.DistributedDataParallel(
+ module=model,
+ device_ids=[self.rank],
+ output_device=self.rank,
+ find_unused_parameters=self.config("find_unused_parameters", False),
+ )
+
+ pytorch_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
+ logger.info(f"total parameters count {pytorch_params} distributed {self.is_distributed}")
+ return model
+
+ def get_train_dataset_data(self):
+ train_files, valid_files = [], []
+ dataset_data = self.config("train#dataset#data")
+ val_key = None
+ if isinstance(dataset_data, dict):
+ val_key = dataset_data.get("key", None)
+ data_list_files = dataset_data["data_list_files"]
+
+ if isinstance(data_list_files, str):
+ data_list_files = ConfigParser.load_config_file(
+ data_list_files
+ ) # if it's a path to a separate file with a list of datasets
+ else:
+ data_list_files = ensure_tuple(data_list_files)
+
+ if self.global_rank == 0:
+ print("Using data_list_files ", data_list_files)
+
+ for idx, d in enumerate(data_list_files):
+ logger.info(f"adding datalist ({idx}): {d['datalist']}")
+ t, v = datafold_read(datalist=d["datalist"], basedir=d["basedir"], fold=self.config("fold"))
+
+ if val_key is not None:
+ v, _ = datafold_read(datalist=d["datalist"], basedir=d["basedir"], fold=-1, key=val_key) # e.g. testing
+
+ for item in t:
+ item["datalist_id"] = idx
+ item["datalist_count"] = len(t)
+ for item in v:
+ item["datalist_id"] = idx
+ item["datalist_count"] = len(v)
+ train_files.extend(t)
+ valid_files.extend(v)
+
+ if self.config("quick", False):
+ logger.info("quick_data")
+ train_files = train_files[:8]
+ valid_files = valid_files[:7]
+ if not valid_files:
+ logger.warning("No validation data found.")
+ return train_files, valid_files
+
+ def read_val_datalists(self, section="validate", data_list_files=None, val_key=None, merge=True):
+ """read the corresponding folds of the datalist for validation or inference"""
+ dataset_data = self.config(f"{section}#dataset#data")
+
+ if isinstance(dataset_data, list):
+ return dataset_data
+
+ if data_list_files is None:
+ data_list_files = dataset_data["data_list_files"]
+
+ if isinstance(data_list_files, str):
+ data_list_files = ConfigParser.load_config_file(
+ data_list_files
+ ) # if it's a path to a separate file with a list of datasets
+ else:
+ data_list_files = ensure_tuple(data_list_files)
+
+ if val_key is None:
+ val_key = dataset_data.get("key", None)
+
+ val_files, idx = [], 0
+ for d in data_list_files:
+ if val_key is not None:
+ v_files, _ = datafold_read(datalist=d["datalist"], basedir=d["basedir"], fold=-1, key=val_key)
+ else:
+ _, v_files = datafold_read(datalist=d["datalist"], basedir=d["basedir"], fold=self.config("fold"))
+ logger.info(f"adding datalist ({idx} -- {val_key}): {d['datalist']} {len(v_files)}")
+ if merge:
+ val_files.extend(v_files)
+ else:
+ val_files.append(v_files)
+ idx += 1
+
+ if self.config("quick", False):
+ logger.info("quick_data")
+ val_files = val_files[:8] if merge else [val_files[0][:8]]
+ return val_files
+
+ def get_train_preprocessing(self):
+ roi_size = self.config("train#dataset#preprocessing#roi_size")
+
+ train_xforms = []
+ train_xforms.append(LoadTiffd(keys=["image", "label"]))
+ train_xforms.append(mt.EnsureTyped(keys=["image", "label"], data_type="tensor", dtype=torch.float))
+ if self.config("prescale", True):
+ print("Prescaling images to 0..1")
+ train_xforms.append(mt.ScaleIntensityd(keys="image", minv=0, maxv=1, channel_wise=True))
+ train_xforms.append(mt.ScaleIntensityd(keys="image", minv=0, maxv=1, channel_wise=True))
+ train_xforms.append(
+ mt.ScaleIntensityRangePercentilesd(
+ keys="image", lower=1, upper=99, b_min=0.0, b_max=1.0, channel_wise=True, clip=True
+ )
+ )
+ train_xforms.append(mt.SpatialPadd(keys=["image", "label"], spatial_size=roi_size))
+ train_xforms.append(
+ mt.RandSpatialCropd(keys=["image", "label"], roi_size=roi_size)
+ ) # crop roi_size (if image is large)
+
+ # # add augmentations
+ train_xforms.extend(
+ [
+ mt.RandAffined(
+ keys=["image", "label"],
+ prob=0.5,
+ rotate_range=np.pi, # from -pi to pi
+ scale_range=[-0.5, 0.5], # from 0.5 to 1.5
+ mode=["bilinear", "nearest"],
+ spatial_size=roi_size,
+ cache_grid=True,
+ padding_mode="border",
+ ),
+ mt.RandAxisFlipd(keys=["image", "label"], prob=0.5),
+ mt.RandGaussianNoised(keys=["image"], prob=0.25, mean=0, std=0.1),
+ mt.RandAdjustContrastd(keys=["image"], prob=0.25, gamma=(1, 2)),
+ mt.RandGaussianSmoothd(keys=["image"], prob=0.25, sigma_x=(1, 2)),
+ mt.RandHistogramShiftd(keys=["image"], prob=0.25, num_control_points=3),
+ mt.RandGaussianSharpend(keys=["image"], prob=0.25),
+ ]
+ )
+
+ train_xforms.append(
+ LabelsToFlows(keys="label", flow_key="flow")
+ ) # finally create new key "flows" with 3 channels 1) foreground 2) dx flow 3) dy flow
+
+ return train_xforms
+
+ def get_val_preprocessing(self):
+ val_xforms = []
+ val_xforms.append(LoadTiffd(keys=["image", "label"], allow_missing_keys=True))
+ val_xforms.append(
+ mt.EnsureTyped(keys=["image", "label"], data_type="tensor", dtype=torch.float, allow_missing_keys=True)
+ )
+
+ if self.config("prescale", True):
+ print("Prescaling val images to 0..1")
+ val_xforms.append(mt.ScaleIntensityd(keys="image", minv=0, maxv=1, channel_wise=True))
+
+ val_xforms.append(
+ mt.ScaleIntensityRangePercentilesd(
+ keys="image", lower=1, upper=99, b_min=0.0, b_max=1.0, channel_wise=True, clip=True
+ )
+ )
+ val_xforms.append(LabelsToFlows(keys="label", flow_key="flow", allow_missing_keys=True))
+
+ return val_xforms
+
+ def get_train_dataset(self):
+ train_dataset_data = self.config("train#dataset#data")
+ if isinstance(train_dataset_data, list): # FIXME, why check
+ train_files = train_dataset_data
+ else:
+ train_files, _ = self.train_dataset_data
+ logger.info(f"train files {len(train_files)}")
+ return Dataset(data=train_files, transform=mt.Compose(self.train_preprocessing))
+
+ def get_val_dataset(self):
+ """this is to be used for validation during training"""
+ val_dataset_data = self.config("validate#dataset#data")
+ if isinstance(val_dataset_data, list): # FIXME, why check
+ valid_files = val_dataset_data
+ else:
+ _, valid_files = self.train_dataset_data
+ return Dataset(data=valid_files, transform=mt.Compose(self.val_preprocessing))
+
+ def set_val_datalist(self, datalist_py):
+ self.parser["validate#dataset#data"] = datalist_py
+ self._props.pop("val_loader", None)
+ self._props.pop("val_dataset", None)
+ self._props.pop("val_sampler", None)
+
+ def get_train_sampler(self):
+ if self.config("use_weighted_sampler", False):
+ data = self.train_dataset.data
+ logger.info(f"Using weighted sampler, first item {data[0]}")
+ sample_weights = 1.0 / torch.as_tensor(
+ [item.get("datalist_count", 1.0) for item in data], dtype=torch.float
+ ) # inverse proportional to sub-datalist count
+ # if we are using weighed sampling, the number of iterations epoch must be provided
+ # (cant use a dataset length anymore)
+ num_samples_per_epoch = self.config("num_samples_per_epoch", None)
+ if num_samples_per_epoch is None:
+ num_samples_per_epoch = len(data) # a workaround if not provided
+ logger.warning(
+ "We are using weighted random sampler, but num_samples_per_epoch is not provided, "
+ f"so using {num_samples_per_epoch} full data length as a workaround!"
+ )
+
+ if self.is_distributed:
+ return DistributedWeightedSampler(
+ self.train_dataset, shuffle=True, weights=sample_weights, num_samples=num_samples_per_epoch
+ ) # custom implementation, as Pytorch does not have one
+ return WeightedRandomSampler(weights=sample_weights, num_samples=num_samples_per_epoch)
+
+ if self.is_distributed:
+ return DistributedSampler(self.train_dataset, shuffle=True)
+ return None
+
+ def get_val_sampler(self):
+ if self.is_distributed:
+ return DistributedSampler(self.val_dataset, shuffle=False)
+ return None
+
+ def get_train_loader(self):
+ sampler = self.train_sampler
+ return DataLoader(
+ self.train_dataset,
+ batch_size=self.config("train#batch_size"),
+ shuffle=(sampler is None),
+ sampler=sampler,
+ pin_memory=True,
+ num_workers=self.config("train#num_workers"),
+ )
+
+ def get_val_loader(self):
+ sampler = self.val_sampler
+ return DataLoader(
+ self.val_dataset,
+ batch_size=self.config("validate#batch_size"),
+ shuffle=False,
+ sampler=sampler,
+ pin_memory=True,
+ num_workers=self.config("validate#num_workers"),
+ )
+
+ def train(self):
+ config = self.config
+ distributed = self.is_distributed
+ sliding_inferrer = config("inferer#sliding_inferer")
+ use_amp = config("amp")
+
+ amp_dtype = {"float32": torch.float32, "bfloat16": torch.bfloat16, "float16": torch.float16}[
+ config("amp_dtype")
+ ]
+ if amp_dtype == torch.bfloat16 and not torch.cuda.is_bf16_supported():
+ amp_dtype = torch.float16
+ logger.warning(
+ "bfloat16 dtype is not support on your device, changing to float16, use --amp_dtype=float16 to set manually"
+ )
+
+ use_gradscaler = use_amp and amp_dtype == torch.float16
+ logger.info(f"Using grad scaler {use_gradscaler} amp_dtype {amp_dtype} use_amp {use_amp}")
+ grad_scaler = GradScaler(enabled=use_gradscaler) # using GradScaler only for AMP float16 (not bfloat16)
+
+ loss_function = config("loss_function")
+ acc_function = config("key_metric")
+
+ ckpt_path = config("ckpt_path")
+ channels_last = config("channels_last")
+
+ num_epochs_per_saving = config("train#trainer#num_epochs_per_saving")
+ num_epochs_per_validation = config("train#trainer#num_epochs_per_validation")
+ num_epochs = config("train#trainer#max_epochs")
+ val_schedule_list = self.schedule_validation_epochs(
+ num_epochs=num_epochs, num_epochs_per_validation=num_epochs_per_validation
+ )
+ logger.info(f"Scheduling validation loops at epochs: {val_schedule_list}")
+
+ train_loader = self.train_loader
+ val_loader = self.val_loader
+ optimizer = config("optimizer")
+ model = self.network
+
+ tb_writer = None
+ csv_path = progress_path = None
+
+ if self.global_rank == 0 and ckpt_path is not None:
+ # rank 0 is responsible for heavy lifting of logging/saving
+ progress_path = os.path.join(ckpt_path, "progress.yaml")
+
+ tb_writer = SummaryWriter(log_dir=ckpt_path)
+ logger.info(f"Writing Tensorboard logs to {tb_writer.log_dir}")
+
+ if mlflow_is_imported:
+ if config("mlflow_tracking_uri", None) is not None:
+ mlflow.set_tracking_uri(config("mlflow_tracking_uri"))
+ mlflow.set_experiment("vista2d")
+
+ mlflow_run_name = config("mlflow_run_name", f'vista2d train fold{config("fold")}')
+ mlflow.start_run(
+ run_name=mlflow_run_name, log_system_metrics=config("mlflow_log_system_metrics", False)
+ )
+ mlflow.log_params(self.parser.config)
+ mlflow.log_dict(self.parser.config, "hyper_parameters.yaml") # experimental
+
+ csv_path = os.path.join(ckpt_path, "accuracy_history.csv")
+ self.save_history_csv(
+ csv_path=csv_path,
+ header=["epoch", "metric", "loss", "iter", "time", "train_time", "validation_time", "epoch_time"],
+ )
+
+ do_torch_save = (
+ (self.global_rank == 0) and ckpt_path and config("ckpt_save") and not config("train#skip", False)
+ )
+ best_ckpt_path = os.path.join(ckpt_path, "model.pt")
+ intermediate_ckpt_path = os.path.join(ckpt_path, "model_final.pt")
+
+ best_metric = float(config("best_metric", -1))
+ start_epoch = config("start_epoch", 0)
+ best_metric_epoch = -1
+ pre_loop_time = time.time()
+ report_num_epochs = num_epochs
+ train_time = validation_time = 0
+ val_acc_history = []
+
+ if start_epoch > 0:
+ val_schedule_list = [v for v in val_schedule_list if v >= start_epoch]
+ if len(val_schedule_list) == 0:
+ val_schedule_list = [start_epoch]
+ print(f"adjusted schedule_list {val_schedule_list}")
+
+ logger.info(
+ f"Using num_epochs => {num_epochs}\n "
+ f"Using start_epoch => {start_epoch}\n "
+ f"batch_size => {config('train#batch_size')} \n "
+ f"num_warmup_epochs => {config('train#trainer#num_warmup_epochs')} \n "
+ )
+
+ lr_scheduler = config("lr_scheduler")
+ if lr_scheduler is not None and start_epoch > 0:
+ lr_scheduler.last_epoch = start_epoch
+
+ range_num_epochs = range(start_epoch, num_epochs)
+
+ if distributed:
+ dist.barrier()
+
+ if self.global_rank == 0 and tb_writer is not None and mlflow_is_imported and mlflow.is_tracking_uri_set():
+ mlflow.log_param("len_train_set", len(train_loader.dataset))
+ mlflow.log_param("len_val_set", len(val_loader.dataset))
+
+ for epoch in range_num_epochs:
+ report_epoch = epoch
+
+ if distributed:
+ if isinstance(train_loader.sampler, DistributedSampler):
+ train_loader.sampler.set_epoch(epoch)
+ dist.barrier()
+
+ epoch_time = start_time = time.time()
+
+ train_loss, train_acc = 0, 0
+
+ if not config("train#skip", False):
+ train_loss, train_acc = self.train_epoch(
+ model=model,
+ train_loader=train_loader,
+ optimizer=optimizer,
+ loss_function=loss_function,
+ acc_function=acc_function,
+ grad_scaler=grad_scaler,
+ epoch=report_epoch,
+ rank=self.rank,
+ global_rank=self.global_rank,
+ num_epochs=report_num_epochs,
+ use_amp=use_amp,
+ amp_dtype=amp_dtype,
+ channels_last=channels_last,
+ device=config("device"),
+ )
+
+ train_time = time.time() - start_time
+ logger.info(
+ f"Latest training {report_epoch}/{report_num_epochs - 1} "
+ f"loss: {train_loss:.4f} time {train_time:.2f}s "
+ f"lr: {optimizer.param_groups[0]['lr']:.4e}"
+ )
+
+ if self.global_rank == 0 and tb_writer is not None:
+ tb_writer.add_scalar("train/loss", train_loss, report_epoch)
+
+ if mlflow_is_imported and mlflow.is_tracking_uri_set():
+ mlflow.log_metric("train/loss", train_loss, step=report_epoch)
+ mlflow.log_metric("train/epoch_time", train_time, step=report_epoch)
+
+ # validate every num_epochs_per_validation epochs (defaults to 1, every epoch)
+ val_acc_mean = -1
+ if (
+ len(val_schedule_list) > 0
+ and epoch + 1 >= val_schedule_list[0]
+ and val_loader is not None
+ and len(val_loader) > 0
+ ):
+ val_schedule_list.pop(0)
+
+ start_time = time.time()
+ torch.cuda.empty_cache()
+
+ val_loss, val_acc = self.val_epoch(
+ model=model,
+ val_loader=val_loader,
+ sliding_inferrer=sliding_inferrer,
+ loss_function=loss_function,
+ acc_function=acc_function,
+ epoch=report_epoch,
+ rank=self.rank,
+ global_rank=self.global_rank,
+ num_epochs=report_num_epochs,
+ use_amp=use_amp,
+ amp_dtype=amp_dtype,
+ channels_last=channels_last,
+ device=config("device"),
+ )
+
+ torch.cuda.empty_cache()
+ validation_time = time.time() - start_time
+
+ val_acc_mean = float(np.mean(val_acc))
+ val_acc_history.append((report_epoch, val_acc_mean))
+
+ if self.global_rank == 0:
+ logger.info(
+ f"Latest validation {report_epoch}/{report_num_epochs - 1} "
+ f"loss: {val_loss:.4f} acc_avg: {val_acc_mean:.4f} acc: {val_acc} time: {validation_time:.2f}s"
+ )
+
+ if tb_writer is not None:
+ tb_writer.add_scalar("val/acc", val_acc_mean, report_epoch)
+ tb_writer.add_scalar("val/loss", val_loss, report_epoch)
+ if mlflow_is_imported and mlflow.is_tracking_uri_set():
+ mlflow.log_metric("val/acc", val_acc_mean, step=report_epoch)
+ mlflow.log_metric("val/epoch_time", validation_time, step=report_epoch)
+
+ timing_dict = {
+ "time": f"{(time.time() - pre_loop_time) / 3600:.2f} hr",
+ "train_time": f"{train_time:.2f}s",
+ "validation_time": f"{validation_time:.2f}s",
+ "epoch_time": f"{time.time() - epoch_time:.2f}s",
+ }
+
+ if val_acc_mean > best_metric:
+ logger.info(f"New best metric ({best_metric:.6f} --> {val_acc_mean:.6f}). ")
+ best_metric, best_metric_epoch = val_acc_mean, report_epoch
+ save_time = 0
+ if do_torch_save:
+ save_time = self.checkpoint_save(
+ ckpt=best_ckpt_path, model=model, epoch=best_metric_epoch, best_metric=best_metric
+ )
+
+ if progress_path is not None:
+ self.save_progress_yaml(
+ progress_path=progress_path,
+ ckpt=best_ckpt_path if do_torch_save else None,
+ best_avg_score_epoch=best_metric_epoch,
+ best_avg_score=best_metric,
+ save_time=save_time,
+ **timing_dict,
+ )
+ if csv_path is not None:
+ self.save_history_csv(
+ csv_path=csv_path,
+ epoch=report_epoch,
+ metric=f"{val_acc_mean:.4f}",
+ loss=f"{train_loss:.4f}",
+ iter=report_epoch * len(train_loader.dataset),
+ **timing_dict,
+ )
+
+ # sanity check
+ if epoch > max(20, num_epochs / 4) and 0 <= val_acc_mean < 0.01 and config("stop_on_lowacc", True):
+ logger.info(
+ f"Accuracy seems very low at epoch {report_epoch}, acc {val_acc_mean}. "
+ "Most likely optimization diverged, try setting a smaller learning_rate"
+ f" than {config('learning_rate')}"
+ )
+ raise ValueError(
+ f"Accuracy seems very low at epoch {report_epoch}, acc {val_acc_mean}. "
+ "Most likely optimization diverged, try setting a smaller learning_rate"
+ f" than {config('learning_rate')}"
+ )
+
+ # save intermediate checkpoint every num_epochs_per_saving epochs
+ if do_torch_save and ((epoch + 1) % num_epochs_per_saving == 0 or (epoch + 1) >= num_epochs):
+ if report_epoch != best_metric_epoch:
+ self.checkpoint_save(
+ ckpt=intermediate_ckpt_path, model=model, epoch=report_epoch, best_metric=val_acc_mean
+ )
+ else:
+ try:
+ shutil.copyfile(best_ckpt_path, intermediate_ckpt_path) # if already saved once
+ except Exception as err:
+ logger.warning(f"error copying {best_ckpt_path} {intermediate_ckpt_path} {err}")
+ pass
+
+ if lr_scheduler is not None:
+ lr_scheduler.step()
+
+ if self.global_rank == 0:
+ # report time estimate
+ time_remaining_estimate = train_time * (num_epochs - epoch)
+ if val_loader is not None and len(val_loader) > 0:
+ if validation_time == 0:
+ validation_time = train_time
+ time_remaining_estimate += validation_time * len(val_schedule_list)
+
+ logger.info(
+ f"Estimated remaining training time for the current model fold {config('fold')} is "
+ f"{time_remaining_estimate/3600:.2f} hr, "
+ f"running time {(time.time() - pre_loop_time)/3600:.2f} hr, "
+ f"est total time {(time.time() - pre_loop_time + time_remaining_estimate)/3600:.2f} hr \n"
+ )
+
+ # end of main epoch loop
+ train_loader = val_loader = optimizer = None
+
+ # optionally validate best checkpoint
+ logger.info(f"Checking to run final testing {config('run_final_testing')}")
+ if config("run_final_testing"):
+ if distributed:
+ dist.barrier()
+ _ckpt_name = best_ckpt_path if os.path.exists(best_ckpt_path) else intermediate_ckpt_path
+ if not os.path.exists(_ckpt_name):
+ logger.info(f"Unable to validate final no checkpoints found {best_ckpt_path}, {intermediate_ckpt_path}")
+ else:
+ # self._props.pop("network", None)
+ # self._set_props.pop("network", None)
+ gc.collect()
+ torch.cuda.empty_cache()
+ best_metric = self.run_final_testing(
+ pretrained_ckpt_path=_ckpt_name,
+ progress_path=progress_path,
+ best_metric_epoch=best_metric_epoch,
+ pre_loop_time=pre_loop_time,
+ )
+
+ if (
+ self.global_rank == 0
+ and tb_writer is not None
+ and mlflow_is_imported
+ and mlflow.is_tracking_uri_set()
+ ):
+ mlflow.log_param("acc_testing", val_acc_mean)
+ mlflow.log_metric("acc_testing", val_acc_mean)
+
+ if tb_writer is not None:
+ tb_writer.flush()
+ tb_writer.close()
+
+ if mlflow_is_imported and mlflow.is_tracking_uri_set():
+ mlflow.end_run()
+
+ logger.info(
+ f"=== DONE: best_metric: {best_metric:.4f} at epoch: {best_metric_epoch} of {report_num_epochs}."
+ f"Training time {(time.time() - pre_loop_time)/3600:.2f} hr."
+ )
+ return best_metric
+
+ def run_final_testing(self, pretrained_ckpt_path, progress_path, best_metric_epoch, pre_loop_time):
+ logger.info("Running final best model testing set!")
+
+ # validate
+ start_time = time.time()
+
+ self._props.pop("network", None)
+ self.parser["pretrained_ckpt_path"] = pretrained_ckpt_path
+ self.parser["validate#evaluator#postprocessing"] = None # not saving images
+
+ val_acc_mean, val_loss, val_acc = self.validate(val_key="testing")
+ validation_time = f"{time.time() - start_time:.2f}s"
+ val_acc_mean = float(np.mean(val_acc))
+ logger.info(f"Testing: loss: {val_loss:.4f} acc_avg: {val_acc_mean:.4f} acc {val_acc} time {validation_time}")
+
+ if self.global_rank == 0 and progress_path is not None:
+ self.save_progress_yaml(
+ progress_path=progress_path,
+ ckpt=pretrained_ckpt_path,
+ best_avg_score_epoch=best_metric_epoch,
+ best_avg_score=val_acc_mean,
+ validation_time=validation_time,
+ run_final_testing=True,
+ time=f"{(time.time() - pre_loop_time) / 3600:.2f} hr",
+ )
+ return val_acc_mean
+
+ def validate(self, validation_files=None, val_key=None, datalist=None):
+ if self.config("pretrained_ckpt_name", None) is None and self.config("pretrained_ckpt_path", None) is None:
+ self.parser["pretrained_ckpt_name"] = "model.pt"
+ logger.info("Using default model.pt checkpoint for validation.")
+
+ grouping = self.config("validate#grouping", False) # whether to computer average per datalist
+ if validation_files is None:
+ validation_files = self.read_val_datalists("validate", datalist, val_key=val_key, merge=not grouping)
+ if len(validation_files) == 0:
+ logger.warning(f"No validation files found {datalist} {val_key}!")
+ return 0, 0, 0
+ if not grouping or not isinstance(validation_files[0], (list, tuple)):
+ validation_files = [validation_files]
+ logger.info(f"validation file groups {len(validation_files)} grouping {grouping}")
+ val_acc_dict = {}
+
+ amp_dtype = {"float32": torch.float32, "bfloat16": torch.bfloat16, "float16": torch.float16}[
+ self.config("amp_dtype")
+ ]
+ if amp_dtype == torch.bfloat16 and not torch.cuda.is_bf16_supported():
+ amp_dtype = torch.float16
+ logger.warning(
+ "bfloat16 dtype is not support on your device, changing to float16, use --amp_dtype=float16 to set manually"
+ )
+
+ for datalist_id, group_files in enumerate(validation_files):
+ self.set_val_datalist(group_files)
+ val_loader = self.val_loader
+
+ start_time = time.time()
+ val_loss, val_acc = self.val_epoch(
+ model=self.network,
+ val_loader=val_loader,
+ sliding_inferrer=self.config("inferer#sliding_inferer"),
+ loss_function=self.config("loss_function"),
+ acc_function=self.config("key_metric"),
+ rank=self.rank,
+ global_rank=self.global_rank,
+ use_amp=self.config("amp"),
+ amp_dtype=amp_dtype,
+ post_transforms=self.config("validate#evaluator#postprocessing"),
+ channels_last=self.config("channels_last"),
+ device=self.config("device"),
+ )
+ val_acc_mean = float(np.mean(val_acc))
+ logger.info(
+ f"Validation {datalist_id} complete, loss_avg: {val_loss:.4f} "
+ f"acc_avg: {val_acc_mean:.4f} acc {val_acc} time {time.time() - start_time:.2f}s"
+ )
+ val_acc_dict[datalist_id] = val_acc_mean
+ for k, v in val_acc_dict.items():
+ logger.info(f"group: {k} => {v:.4f}")
+ val_acc_mean = sum(val_acc_dict.values()) / len(val_acc_dict.values())
+ logger.info(f"Testing group score average: {val_acc_mean:.4f}")
+ return val_acc_mean, val_loss, val_acc
+
+ def infer(self, infer_files=None, infer_key=None, datalist=None):
+ if self.config("pretrained_ckpt_name", None) is None and self.config("pretrained_ckpt_path", None) is None:
+ self.parser["pretrained_ckpt_name"] = "model.pt"
+ logger.info("Using default model.pt checkpoint for inference.")
+
+ if infer_files is None:
+ infer_files = self.read_val_datalists("infer", datalist, val_key=infer_key, merge=True)
+ if len(infer_files) == 0:
+ logger.warning(f"no file to infer {datalist} {infer_key}.")
+ return
+ logger.info(f"inference files {len(infer_files)}")
+ self.set_val_datalist(infer_files)
+ val_loader = self.val_loader
+
+ amp_dtype = {"float32": torch.float32, "bfloat16": torch.bfloat16, "float16": torch.float16}[
+ self.config("amp_dtype")
+ ]
+ if amp_dtype == torch.bfloat16 and not torch.cuda.is_bf16_supported():
+ amp_dtype = torch.bfloat16
+ logger.warning(
+ "bfloat16 dtype is not support on your device, changing to float16, use --amp_dtype=float16 to set manually"
+ )
+
+ start_time = time.time()
+ self.val_epoch(
+ model=self.network,
+ val_loader=val_loader,
+ sliding_inferrer=self.config("inferer#sliding_inferer"),
+ loss_function=None,
+ acc_function=None,
+ rank=self.rank,
+ global_rank=self.global_rank,
+ use_amp=self.config("amp"),
+ amp_dtype=amp_dtype,
+ post_transforms=self.config("infer#evaluator#postprocessing"),
+ channels_last=self.config("channels_last"),
+ device=self.config("device"),
+ )
+ logger.info(f"Inference complete time {time.time() - start_time:.2f}s")
+ return
+
+ @torch.no_grad()
+ def val_epoch(
+ self,
+ model,
+ val_loader,
+ sliding_inferrer,
+ loss_function=None,
+ acc_function=None,
+ epoch=0,
+ rank=0,
+ global_rank=0,
+ num_epochs=0,
+ use_amp=True,
+ amp_dtype=torch.float16,
+ post_transforms=None,
+ channels_last=False,
+ device=None,
+ ):
+ model.eval()
+ distributed = dist.is_available() and dist.is_initialized()
+ memory_format = torch.channels_last if channels_last else torch.preserve_format
+
+ run_loss = CumulativeAverage()
+ run_acc = CumulativeAverage()
+ run_loss.append(torch.tensor(0, device=device), count=0)
+
+ avg_loss = avg_acc = 0
+ start_time = time.time()
+
+ # In DDP, each replica has a subset of data, but if total data length is not evenly divisible by num_replicas,
+ # then some replicas has 1 extra repeated item.
+ # For proper validation with batch of 1, we only want to collect metrics for non-repeated items,
+ # hence let's compute a proper subset length
+ nonrepeated_data_length = len(val_loader.dataset)
+ sampler = val_loader.sampler
+ if distributed and isinstance(sampler, DistributedSampler) and not sampler.drop_last:
+ nonrepeated_data_length = len(range(sampler.rank, len(sampler.dataset), sampler.num_replicas))
+
+ for idx, batch_data in enumerate(val_loader):
+ data = batch_data["image"].as_subclass(torch.Tensor).to(memory_format=memory_format, device=device)
+ filename = batch_data["image"].meta[ImageMetaKey.FILENAME_OR_OBJ]
+ batch_size = data.shape[0]
+ loss = acc = None
+
+ with autocast(enabled=use_amp, dtype=amp_dtype):
+ logits = sliding_inferrer(inputs=data, network=model)
+ data = None
+
+ # calc loss
+ if loss_function is not None:
+ target = batch_data["flow"].as_subclass(torch.Tensor).to(device=logits.device)
+ loss = loss_function(logits, target)
+ run_loss.append(loss.to(device=device), count=batch_size)
+ target = None
+
+ pred_mask_all = []
+
+ for b_ind in range(logits.shape[0]): # go over batch dim
+ pred_mask, p = LogitsToLabels()(logits=logits[b_ind], filename=filename)
+ pred_mask_all.append(pred_mask)
+
+ if acc_function is not None:
+ label = batch_data["label"].as_subclass(torch.Tensor)
+
+ for b_ind in range(label.shape[0]):
+ acc = acc_function(pred_mask_all[b_ind], label[b_ind, 0].long())
+ acc = acc.detach().clone() if isinstance(acc, torch.Tensor) else torch.tensor(acc)
+
+ if idx < nonrepeated_data_length:
+ run_acc.append(acc.to(device=device), count=1)
+ else:
+ run_acc.append(torch.zeros_like(acc, device=device), count=0)
+ label = None
+
+ avg_loss = loss.cpu() if loss is not None else 0
+ avg_acc = acc.cpu().numpy() if acc is not None else 0
+
+ logger.info(
+ f"Val {epoch}/{num_epochs} {idx}/{len(val_loader)} "
+ f"loss: {avg_loss:.4f} acc {avg_acc} time {time.time() - start_time:.2f}s"
+ )
+
+ if post_transforms:
+ seg = torch.from_numpy(np.stack(pred_mask_all, axis=0).astype(np.int32)).unsqueeze(1)
+ batch_data["seg"] = convert_to_dst_type(
+ seg, batch_data["image"], dtype=torch.int32, device=torch.device("cpu")
+ )[0]
+ for bd in decollate_batch(batch_data):
+ post_transforms(bd) # (currently only to save output mask)
+
+ start_time = time.time()
+
+ label = target = data = batch_data = None
+
+ if distributed:
+ dist.barrier()
+
+ avg_loss = run_loss.aggregate()
+ avg_acc = run_acc.aggregate()
+
+ if np.any(avg_acc < 0):
+ dist.barrier()
+ logger.warning(f"Avg accuracy is negative ({avg_acc}), something went wrong!!!!!")
+
+ return avg_loss, avg_acc
+
+ def train_epoch(
+ self,
+ model,
+ train_loader,
+ optimizer,
+ loss_function,
+ acc_function,
+ grad_scaler,
+ epoch,
+ rank,
+ global_rank=0,
+ num_epochs=0,
+ use_amp=True,
+ amp_dtype=torch.float16,
+ channels_last=False,
+ device=None,
+ ):
+ model.train()
+ memory_format = torch.channels_last if channels_last else torch.preserve_format
+
+ run_loss = CumulativeAverage()
+
+ start_time = time.time()
+ avg_loss = avg_acc = 0
+ for idx, batch_data in enumerate(train_loader):
+ data = batch_data["image"].as_subclass(torch.Tensor).to(memory_format=memory_format, device=device)
+ target = batch_data["flow"].as_subclass(torch.Tensor).to(memory_format=memory_format, device=device)
+
+ optimizer.zero_grad(set_to_none=True)
+
+ with autocast(enabled=use_amp, dtype=amp_dtype):
+ logits = model(data)
+
+ # print('logits', logits.shape, logits.dtype)
+ loss = loss_function(logits.float(), target)
+
+ grad_scaler.scale(loss).backward()
+ grad_scaler.step(optimizer)
+ grad_scaler.update()
+
+ batch_size = data.shape[0]
+
+ run_loss.append(loss, count=batch_size)
+ avg_loss = run_loss.aggregate()
+
+ logger.info(
+ f"Epoch {epoch}/{num_epochs} {idx}/{len(train_loader)} "
+ f"loss: {avg_loss:.4f} time {time.time() - start_time:.2f}s "
+ )
+ start_time = time.time()
+
+ optimizer.zero_grad(set_to_none=True)
+
+ data = None
+ target = None
+ batch_data = None
+
+ return avg_loss, avg_acc
+
+ def save_history_csv(self, csv_path=None, header=None, **kwargs):
+ if csv_path is not None:
+ if header is not None:
+ with open(csv_path, "a") as myfile:
+ wrtr = csv.writer(myfile, delimiter="\t")
+ wrtr.writerow(header)
+ if len(kwargs):
+ with open(csv_path, "a") as myfile:
+ wrtr = csv.writer(myfile, delimiter="\t")
+ wrtr.writerow(list(kwargs.values()))
+
+ def save_progress_yaml(self, progress_path=None, ckpt=None, **report):
+ if ckpt is not None:
+ report["model"] = ckpt
+
+ report["date"] = str(datetime.now())[:19]
+
+ if progress_path is not None:
+ yaml.add_representer(
+ float, lambda dumper, value: dumper.represent_scalar("tag:yaml.org,2002:float", f"{value:.4f}")
+ )
+ with open(progress_path, "a") as progress_file:
+ yaml.dump([report], stream=progress_file, allow_unicode=True, default_flow_style=None, sort_keys=False)
+
+ logger.info("Progress:" + ",".join(f" {k}: {v}" for k, v in report.items()))
+
+ def checkpoint_save(self, ckpt: str, model: torch.nn.Module, **kwargs):
+ # save checkpoint and config
+ save_time = time.time()
+ if isinstance(model, torch.nn.parallel.DistributedDataParallel):
+ state_dict = model.module.state_dict()
+ else:
+ state_dict = model.state_dict()
+
+ if self.config("compile", False):
+ # remove key prefix of compiled models
+ state_dict = OrderedDict(
+ (k[len("_orig_mod.") :] if k.startswith("_orig_mod.") else k, v) for k, v in state_dict.items()
+ )
+
+ torch.save({"state_dict": state_dict, "config": self.parser.config, **kwargs}, ckpt)
+
+ save_time = time.time() - save_time
+ logger.info(f"Saving checkpoint process: {ckpt}, {kwargs}, save_time {save_time:.2f}s")
+
+ return save_time
+
+ def checkpoint_load(self, ckpt: str, model: torch.nn.Module, **kwargs):
+ # load checkpoint
+ if not os.path.isfile(ckpt):
+ logger.warning("Invalid checkpoint file: " + str(ckpt))
+ return
+ checkpoint = torch.load(ckpt, map_location="cpu")
+
+ model.load_state_dict(checkpoint["state_dict"], strict=True)
+ epoch = checkpoint.get("epoch", 0)
+ best_metric = checkpoint.get("best_metric", 0)
+
+ if self.config("continue", False):
+ if "epoch" in checkpoint:
+ self.parser["start_epoch"] = checkpoint["epoch"]
+ if "best_metric" in checkpoint:
+ self.parser["best_metric"] = checkpoint["best_metric"]
+
+ logger.info(
+ f"=> loaded checkpoint {ckpt} (epoch {epoch}) "
+ f"(best_metric {best_metric}) setting start_epoch {self.config('start_epoch')}"
+ )
+ self.parser["start_epoch"] = int(self.config("start_epoch")) + 1
+ return
+
+ def schedule_validation_epochs(self, num_epochs, num_epochs_per_validation=None, fraction=0.16) -> list:
+ """
+ Schedule of epochs to validate (progressively more frequently)
+ num_epochs - total number of epochs
+ num_epochs_per_validation - if provided use a linear schedule with this step
+ init_step
+ """
+
+ if num_epochs_per_validation is None:
+ x = (np.sin(np.linspace(0, np.pi / 2, max(10, int(fraction * num_epochs)))) * num_epochs).astype(int)
+ x = np.cumsum(np.sort(np.diff(np.unique(x)))[::-1])
+ x[-1] = num_epochs
+ x = x.tolist()
+ else:
+ if num_epochs_per_validation >= num_epochs:
+ x = [num_epochs_per_validation]
+ else:
+ x = list(range(num_epochs_per_validation, num_epochs, num_epochs_per_validation))
+
+ if len(x) == 0:
+ x = [0]
+
+ return x
+
+
+def main(**kwargs) -> None:
+ workflow = VistaCell(**kwargs)
+ workflow.initialize()
+ workflow.run()
+ workflow.finalize()
+
+
+if __name__ == "__main__":
+ # to be able to run directly as python scripts/workflow.py --config_file=...
+ # for debugging and development
+
+ from pathlib import Path
+
+ sys.path.append(str(Path(__file__).parent.parent))
+
+ # from scripts import *
+
+ fire, fire_is_imported = optional_import("fire")
+ if fire_is_imported:
+ fire.Fire(main)
+ else:
+ print("Missing package: fire")


