Bar plots
Violin plots
Why do we focus on these common plots? Our eyes are better at distinguishing certain visual features more than others. All of these plots are focused on their position to depict data, which gives us the most effective visual scale.
Let’s load in our genomics datasets and start making some plots from them.
@@ -363,9 +364,13 @@5.4 Basic plot customization## <string>:1: UserWarning: The palette list has more values (6) than needed (3), which may not be intended.

-
-5.5 Exercises
-Exercise for week 5 can be found here.
+
+5.5 Other resources
+We recommend checking out the workshop Better Plots, which showcase examples of how to clean up your plots for clearer communication.
+
+
+5.6 Exercises
+Exercise for week 5 can be found here.
diff --git a/docs/no_toc/data-wrangling-part-1.html b/docs/no_toc/data-wrangling-part-1.html
index 0ece4cc..8f96593 100644
--- a/docs/no_toc/data-wrangling-part-1.html
+++ b/docs/no_toc/data-wrangling-part-1.html
@@ -206,7 +206,8 @@
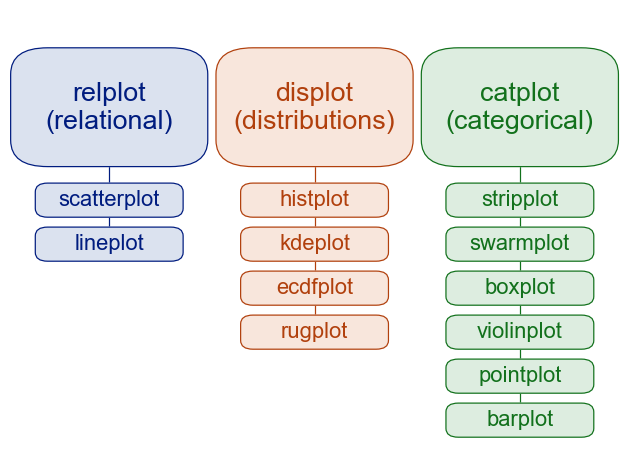
5.2 Relational (between 2 continuous variables)
5.3 Categorical (between 1 categorical and 1 continuous variable)
5.4 Basic plot customization
-5.5 Exercises
+5.5 Other resources
+5.6 Exercises
About the Authors
6 References
@@ -315,10 +316,10 @@ 3.2 Our working Tidy Data: DepMap
## [5 rows x 536 columns]

5.5 Exercises
-Exercise for week 5 can be found here.
+5.5 Other resources
+We recommend checking out the workshop Better Plots, which showcase examples of how to clean up your plots for clearer communication.
+5.6 Exercises
+Exercise for week 5 can be found here.
](https://www.oreilly.com/api/v2/epubs/9781466508910/files/image/fig5-1.png)