-
Notifications
You must be signed in to change notification settings - Fork 4
clusters
baptiste edited this page Jun 22, 2014
·
6 revisions
baptiste Auguié -- 22 June, 2014
A cluster of particles is defined in 3 dimensions by a list with 3 components: positions of the dipoles; Euler angles to orient the polarizability tensor in space; and sizes of the particles (3 semi-axes for ellipsoids). Functions are provided for a few types of common clusters, as well as for their visualisation in 3D using OpenGl or povray.
cl1 <- list(r = rbind(c(0, 0, 0),
c(0, 500, 0)),
angles = rbind(c(0, 0, 0),
c(0, pi/2, pi/3)),
sizes = rbind(c(50, 20, 20),
c(40, 30, 30)))
rgl.ellipsoids(cl1$r, cl1$sizes, cl1$angles, col="gold")
rgl.viewpoint( theta = 0, phi = 20, fov = 70, zoom = 1)
cl2 <- cluster_dimer(d=100,
dihedral=45*pi/180, alpha1=10*pi/180, alpha2=0,
a=35, b=12)
rgl.ellipsoids(cl2$r, cl2$sizes, cl2$angles, col="gold")
cl2 <- cluster_chain(10, pitch=100, a=50, b=30)
rgl.ellipsoids(cl2$r, cl2$sizes, cl2$angles, col="gold")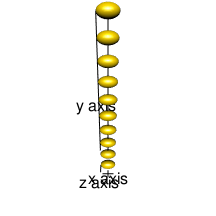
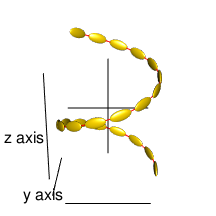
cl3 <- cluster_helix(N=20, R0=500, pitch=1000,
delta=pi/7, delta0=0, right=TRUE,
a=100, b=50, c=20,
angles="helix")
hel3 <- helix(N = 20, R0 = 500, pitch = 1000, delta = pi/7,
delta0 = 0, right = TRUE)
rgl.ellipsoids(cl3$r, cl3$sizes, cl3$angles, col="gold")