-
-
Notifications
You must be signed in to change notification settings - Fork 1.9k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
allow plotly.js to accept numpy buffers #1784
Comments
|
Interesting, definitely seems doable, but is the numpy buffer format accessible from other languages? It looks like a fairly straightforward encoding, so seems like even if it's not natively available elsewhere we could likely generate it - hopefully just translating headers from whatever is natively available. |
|
Here's an R package for writing numpy buffers. |
|
Referencing #860 |
|
Very loosely related, only because Mikola wrote so much useful stuff that's not fully utilized: scijs/ndarray#18 There's been talk of letting the scijs/ndarray constructor accept a plain object with format {data: [...], shape: [...], stride: [...], offset: ...}. It's a bit annoying/inefficient to pack and unpack those via ndarray-unpack and ndarray-pack, but it's pretty trivial. Like if a numpy buffer could get unpacked into an scijs ndarray and then sent to plotly as an array of arrays via |
|
Let me add my 2c on this. Let me repeat the stripped version here: def array_to_json(ar, obj=None):
return {'buffer':memoryview(ar), 'dtype':str(ar.dtype), 'shape':ar.shape}On a lower level is how to send the json (or our extension of json) over the wire. In ipywidgets we allow for memoryview objects (basically a binary blob, we loosly refer to it as a buffer) to be present in the json, which is off course not real valid json, lets call it jsonb. This 'jsonb' then gets split into a real json part, and list of buffers, and a list of 'paths' where these buffers resided in the json structure, on the python side that happens here. The real json part, and the buffers are then send over the wire (websocket) as 1 binary blob, and at the JS side deserialized, most of the js magic is here Let me repeat the example splitting of the jsonb in json, paths and buffers here: >>> state = {'plain': [0, 'text'], 'x': {'ar': memoryview(ar1)}, 'y': {'shape': (10,10), 'data': memoryview(ar2)}}
>>> _remove_buffers(state)
({'plain': [0, 'text']}, {'x': {}, 'y': {'shape': (10, 10)}}, [['x', 'ar'], ['y', 'data']],
[<memory at 0x107ffec48>, <memory at 0x107ffed08>])This can be seen as an extension of json, and I think this part deserves it's own library, which I think can be useful for many other projects. For instance I noticed that bokeh (cc @bryevdv) also has binary transfer on their wish list, so maybe some coordination is useful. Having a jsonb library for python, js, R and c++ would be of interest of many more people I think, beyond ipywidgets, plotly and bokeh. What to do with the buffer object on the js side is is I think up to the app developer, in ipyvolume I now mostly directly use typed arrays (such as Float32Array), and for multi-d cases ndarray. I do however check on the Python side that the array is 'C_CONTIGUOUS', so I do not have to worry about strides. (cc @SylvainCorlay @jasongrout ) PS: @jackparmer I don't transfer the full numpy array data any more (that was before ipywidgets 7), I now serialize only the array data, and send the dtype, and shape separately, i need to remove that code. |
|
Thanks @maartenbreddels for these tips - extremely helpful! We're in the middle of a few other plotly.js projects right now, but are planning to circle back on this in a few weeks. @SylvainCorlay @jasongrout @bryevdv happy to think about standalone implementations for this that could be universally useful. Feel free to chime in if you think of ideas 🥂 |
|
I haven't yet looked into the awesome details here, but we have an older conversation with a similar overall goal in mind (though maybe different context): plotly/plotly.py#550 (comment) At the time we pondered that maybe the Python side could serialize with np.ndarray.tobytes into a WebSocket of |
|
Hi, There is no character based approach what I describe, maybe it seem that way since it is (partly) json, but all the array data is binary transfer with minimal amount of copies. Actually, I wouldn't recommend using cheers, Maarten |
|
Thanks for the clarification @maartenbreddels - your approach looks like the one to be followed! |
|
Copying from @jmmease's #2388 (comment), a proposal on how to encode large typed array inside JSONs:
|
|
Somewhat related to this thread and the topic of data serialization: I came across Apache Arrow which is a cross-language in-memory representation for columnar data to go from the current inefficient copy & convert: |
|
cc @catherinezucker - This came up at gluecon, would be really useful for volume rendering. |
|
This issue has been tagged with A community PR for this feature would certainly be welcome, but our experience is deeper features like this are difficult to complete without the Plotly maintainers leading the effort. Sponsorship range: $10k-$15k What Sponsorship includes:
Please include the link to this issue when contacting us to discuss. |
I saw that |
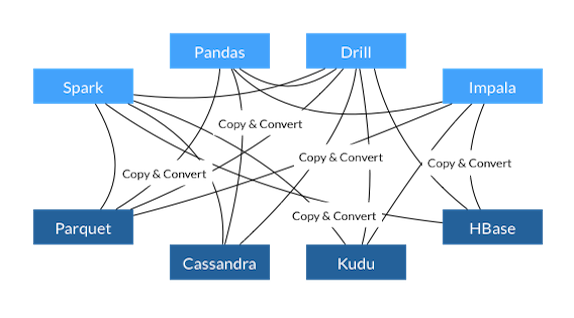
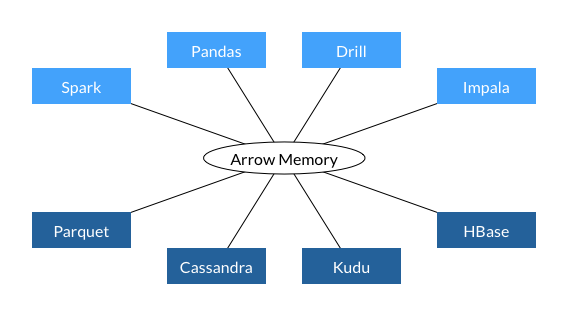
If we allowed all GL plot types to accept numpy buffers for plot x/y/z data, then the Python library could optionally avoid JSON serialization of array data, like @maartenbreddels does in his brillant project ipyvolume:
https://github.com/maartenbreddels/ipyvolume/blob/master/ipyvolume/serialize.py#L95
Here is the deserialization on the JS side:
https://github.com/maartenbreddels/ipyvolume/blob/master/js/src/serialize.js#L16
Plotly.js has all of these incredible WebGL figures for scientific computing, but their potential in Python, R, MATLAB, etc is limited by the JSON serialization step.
In a similar vein, it seems like all GL types should be able to accept Float32Array's directly instead of untyped JS arrays. Currently, it looks like only the Plotly trace type
pointcloudaccepts Float32Array's:https://codepen.io/plotly/pen/GEoPgv
The text was updated successfully, but these errors were encountered: