-
Notifications
You must be signed in to change notification settings - Fork 0
01. Introduction
Sebastian Gregoricchio edited this page Oct 28, 2023
·
3 revisions
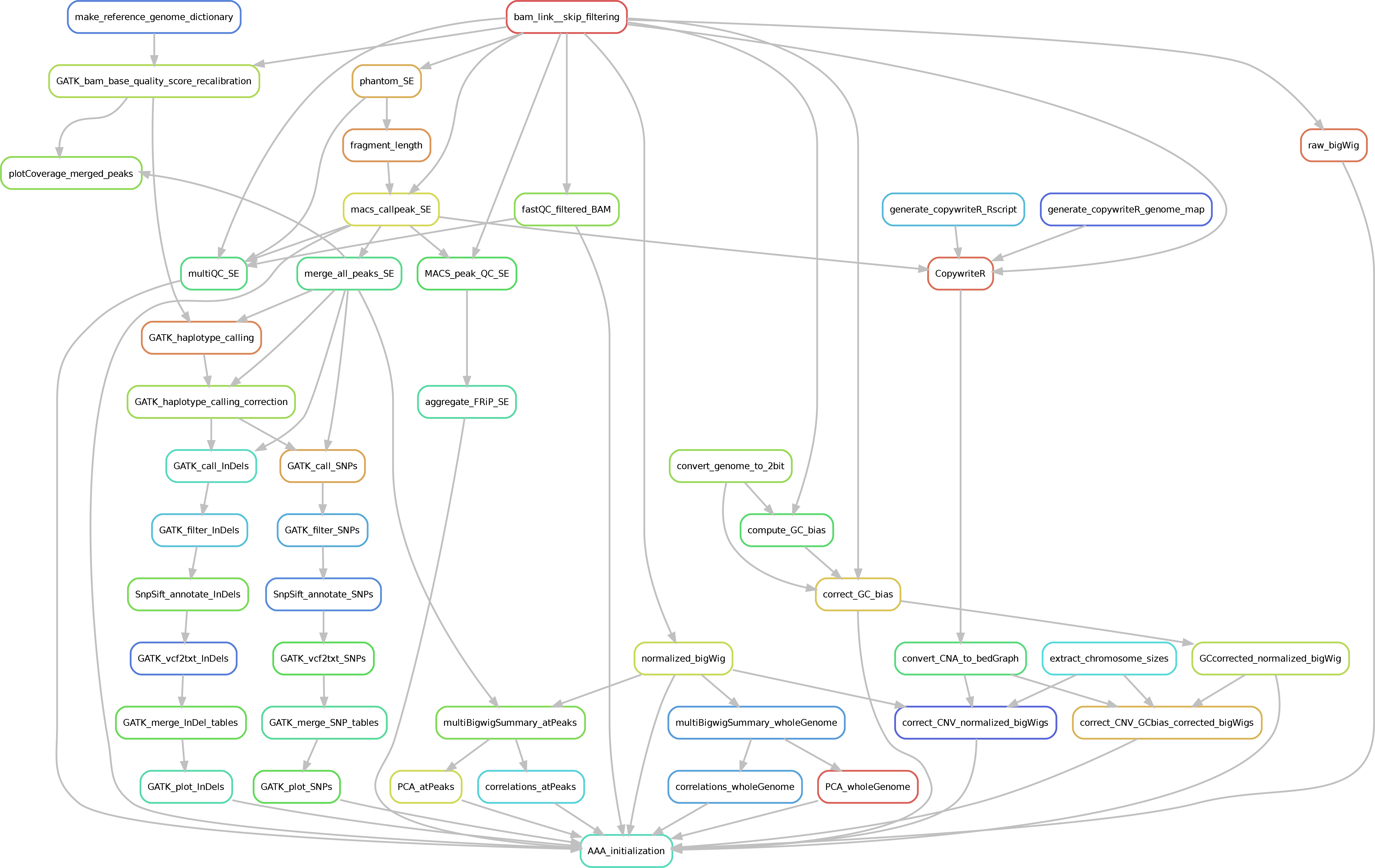
SPACCa is a Snakemake-base Pipeline for the Analyses of ChIP-seq data in Cancer samples. It allows the mapping of fastq files (both paired- and single-end) as well as other downstream analyses starting from the bam files (i.e., mapping filtering, sample correlation, peak calling). The latter, include data normalization, peak calling, GC-bias correction, Copy Number Variation (CNV) detection and signal correction and, identification of SNP/InDel at ChIP-seq peaks.
Fun fact: in italian slang when something "spacca" it means that it rocks.
If you use this pipeline, please cite:
No publication associated yet. "SPACCa: a Snakemake Pipeline for the analyses of ChIP-seq data in Cancer samples.
XYZ, Volume X, Issue X, 2023, XYZ
DOI: XYZ
XYZ, Volume X, Issue X, 2023, XYZ
DOI: XYZ
Contributors