-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathnhs-r-conf_21-fp_workshop.html
1684 lines (1261 loc) · 41.5 KB
/
nhs-r-conf_21-fp_workshop.html
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
816
817
818
819
820
821
822
823
824
825
826
827
828
829
830
831
832
833
834
835
836
837
838
839
840
841
842
843
844
845
846
847
848
849
850
851
852
853
854
855
856
857
858
859
860
861
862
863
864
865
866
867
868
869
870
871
872
873
874
875
876
877
878
879
880
881
882
883
884
885
886
887
888
889
890
891
892
893
894
895
896
897
898
899
900
901
902
903
904
905
906
907
908
909
910
911
912
913
914
915
916
917
918
919
920
921
922
923
924
925
926
927
928
929
930
931
932
933
934
935
936
937
938
939
940
941
942
943
944
945
946
947
948
949
950
951
952
953
954
955
956
957
958
959
960
961
962
963
964
965
966
967
968
969
970
971
972
973
974
975
976
977
978
979
980
981
982
983
984
985
986
987
988
989
990
991
992
993
994
995
996
997
998
999
1000
<!DOCTYPE html>
<html lang="" xml:lang="">
<head>
<title>Functional Programming with R</title>
<meta charset="utf-8" />
<meta name="author" content="[Tom Jemmett][email] | Senior Healthcare Analyst" />
<script src="libs/header-attrs-2.11/header-attrs.js"></script>
<link href="libs/remark-css-0.0.1/default.css" rel="stylesheet" />
<link href="libs/tile-view-0.2.6/tile-view.css" rel="stylesheet" />
<script src="libs/tile-view-0.2.6/tile-view.js"></script>
<link href="libs/animate.css-3.7.2/animate.xaringan.css" rel="stylesheet" />
<link href="libs/animate.css-xaringan-3.7.2/animate.slide_left.css" rel="stylesheet" />
<link href="libs/panelset-0.2.6/panelset.css" rel="stylesheet" />
<script src="libs/panelset-0.2.6/panelset.js"></script>
<link rel="stylesheet" href="css/nhsr.css" type="text/css" />
<link rel="stylesheet" href="css/nhsr-fonts.css" type="text/css" />
</head>
<body>
<textarea id="source">
class: title-slide, left, bottom
# Functional Programming with R
----
## **NHS-R Conference 2021**
### **[Tom Jemmett][email]** | Senior Healthcare Analyst
### **[The Strategy Unit][su]** | Midlands and Lancashire CSU
---
# Session Outline
* What are functions?
* Why do we want to write functions?
* What is functional programming?
* A run through of the `{purrr}` package
- `compose` and `partial`
- `map` and it's variants
- `reduce`
- `safely` and `possibly`
- `keep` and `discard`
- `some`/`every`/`none`
* A brief introduction to Parallel Computation with `{furrr}`
---
# About The Strategy Unit / Me
.pull-left[
"Leading research, analysis and change from within the NHS"
The Strategy Unit is a specialist NHS team, based in Midlands and Lancashire Commissioning Support Unit. We focus on
the application of high-quality, multi-disciplinary analytical work.
Our team comes from diverse backgrounds. Our academic qualifications include maths, economics, history, natural
sciences, medicine, sociology, business and management, psychology and political science. Our career and personal
histories are just as varied.
Our staff are NHS employees, animated by NHS values. The Strategy Unit covers all its costs through project funding.
But this is driven by need, not what we can sell. Any surplus is recycled for public benefit.
[strategyunitwm.nhs.uk](https://strategyunitwm.nhs.uk/)/[GitHub](https://github.com/The-Strategy-Unit)
]
.pull-right[
**Tom Jemmett**
*Senior Healthcare Analyst*
[thomas.jemmett@nhs.net](mailto:thomas.jemmett@nhs.net)
- 10+ years experience within the NHS as a data analyst
- BSc Computer Science and Pure Mathematics (Open University)
- MBCS/AMIMA
- Active member of NHS-R community
- Senior Fellow of NHS-R academy
- @tomjemmett [Twitter](https://twitter.com/tomjemmett)/[GitHub](https://github.com/tomjemmett)
]
---
class: middle, center
# What are functions?
---
## What are functions?
.pull-left[
A function is a process which takes some input, called **arguments**, and produces some output called a **return
value**. Functions may serve the following purposes:
* **Mapping**: Produce some output based on given inputs. A function **maps** input values to output values.
* **Procedures**: A function may be called to perform a sequence of steps. The sequence is known as a procedure, and
programming in this style is known as **procedural programming**.
* **I/O**: Some functions exist to communicate with other parts of the system, such as the screen, storage, system logs
or network.
source: ["What is a pure function?"][mji_what_pure_fn]
]
.pull-right[
``` r
# mapping
fn <- function(x) {
x * x + 1
}
```
``` r
# procedure
counter <- 0
stack <- list()
push <- function(x) {
counter <<- counter + 1
stack[[counter]] <<- x
}
```
``` r
# i/o:
read_csv(filename)
```
]
---
## Vectors in R
In R we have a number of different types of vectors:
* Atomic vectors of 1-dimension, e.g. `c(1, 2, 3)` and `c("hello", "world")`. All values in these vectors contain the
same type of data (they are **homogeneous**)
* Lists are vectors where each item can be an atomic vector, or another list. The items in a list need not be the same
type, nor do they have to be the same length (they are **heterogeneous**). E.g., we can have
``` r
list(
c(1, 2, 3),
c("hello", "world"),
list('a', 'b')
)
```
* Items in atomic vectors or lists can be named, e.g. `c('a' = 1, 'b' = 2)`.
* Dataframes are just special cases of lists: each item in the list must have the same length. Each item is a column,
and the name of the item is the name of the column. That is, `length(df) == ncol(df)`.
* Matrices/Arrays are 2-d and n-d atomic vectors. We won't cover these today, but care needs to be taken when using
these as they can easily be coerced into a 1-d vector (e.g. with a matrix by running through all the items in the first
column, then all the items in the second column etc.)
---
## Environments
.pull-left[
In R, we have the global environment. This is where all variables are created when you assign (`<-`) something in the
console.
When a function is evaluated, it creates it's own environment. All of the arguments that are passed to the function,
along with any variables created in the function are stored in this new environment.
The function's environment's parent will be the global environment, so we can see all of the variables created in the
global environment. Variables that are created in the function's environment aren't visible from the global environment
though.
If we reassign a variable in a function it will take a copy of that variable rather than mutating the value in the
global environment. If we want to update `x` in the global environment we need to use the `<<-` operator.
]
.pull-right[
```r
x <- 1
fn <- function(y) {
x <- x * 2
z <- x + y
z
}
result <- fn(2)
```
```r
exists("z")
```
```
## [1] FALSE
```
```r
x
```
```
## [1] 1
```
]
---
# Why do we want to write functions?
Consider the following code (from the book [R4DS][r4ds]). Can you spot the mistake?
``` r
df <- tibble::tibble(
a = rnorm(10),
b = rnorm(10),
c = rnorm(10),
d = rnorm(10)
)
df$a <- (df$a - min(df$a, na.rm = T)) /
(max(df$a, na.rm = T) - min(df$a, na.rm = T))
df$b <- (df$b - min(df$b, na.rm = T)) /
(max(df$b, na.rm = T) - min(df$a, na.rm = T))
df$c <- (df$c - min(df$c, na.rm = TRUE)) /
(max(df$c, na.rm = T) - min(df$c, na.rm = T))
df$d <- (df$d - min(df$d, na.rm = TRUE)) /
(max(df$d, na.rm = T) - min(df$d, na.rm = T))
```
---
## The mistake
``` r
df$b <- (df$b - min(df$b, na.rm = T)) /
(max(df$b, na.rm = T) - min(df$a, na.rm = T))
```
Often, when we copy and paste code we introduce subtle bugs, in the previous example we forgot to update the one
argument: me call `min(df$a)` rather than `min(df$b)`.
Writing functions can reduce these types of errors by abstracting away the underlying logic.
---
## Creating a function to solve the last problem
.pull-left[
To the right is one way to turn the previous example into a function.
We pass in a numerical vector (`x` to the function), calculate the minimum and maximum values, then rescale the vector.
Finally we update each column, one by one, using this new function.
We still have an issue here with the potential for copy-paste bugs in that we are doing the same thing 4 times, just
changing the column in the data frame that we are using.
We could use a loop, but we will see how functional programming can help us solve this problem more elegantly later.
One important principle functions help us achieve is **DRY** (don't repeat yourself).
]
.pull-right[
``` r
rescale_01 <- function(x) {
min_x <- min(x, na.rm = TRUE)
max_x <- max(x, na.rm = TRUE)
(x - min_x) / (max_x - min_x)
}
```
``` r
# update the columns
df$a <- rescale_01(df$a)
df$b <- rescale_01(df$b)
df$c <- rescale_01(df$c)
df$d <- rescale_01(df$d)
```
``` r
# using a loop
for (i in colnames(df)) {
df[[i]] <- rescale_01(df[[i]])
}
```
]
---
# What is functional programming?
> Functional programming (often abbreviated FP) is the process of building software by composing **pure functions**,
> avoiding **shared state**, **mutable data**, and **side-effects**. Functional programming is **declarative** rather
> than **imperative**, and application state flows through pure functions. Contrast with object oriented programming,
> where application state is usually shared and colocated with methods in objects.
>
> …
>
> Functional code tends to be more concise, more predictable, and easier to test than imperative or object oriented code
> but if you’re unfamiliar with it and the common patterns associated with it, functional code can also seem a lot
> more dense, and the related literature can be impenetrable to newcomers.
["What is functional programming?", Mastering the JavaScript interview][mji_what_fp]
---
## Declarative vs Imperative
.pull-left[
Imperative programming uses statements to change a programs state. It often looks like a series of steps. You are
telling the computer what to do at each step.
``` r
# Correlation panel
panel.cor <- function(x, y){
usr <- par("usr"); on.exit(par(usr))
par(usr = c(0, 1, 0, 1))
r <- round(cor(x, y), digits=2)
txt <- paste0("R = ", r)
cex.cor <- 0.8/strwidth(txt)
text(0.5, 0.5, txt, cex = cex.cor * r)
}
# Customize upper panel
upper.panel<-function(x, y){
points(x,y, pch = 19, col = my_cols[iris$Species])
}
# Create the plots
pairs(iris[,1:4],
lower.panel = panel.cor,
upper.panel = upper.panel)
```
[Source](http://www.sthda.com/english/wiki/scatter-plot-matrices-r-base-graphs)
]
.pull-right[
In contrast, declarative programming focuses on what the program should do, rather than how to do it. Examples of
declarative programming languages include Sql; you do not care how to actually perform the query, you just instruct
the computer what things you want:
``` sql
SELECT
things
FROM
my_table
WHERE
stuff = '...';
```
]
---
## Pure functions
.pull-left[
Pure functions are functions which:
* always produce the same result given the same input
* have no side effects (e.g. reading from a database, writing a file to disk)
* do not use global state (e.g. using variables declared outside of the function)
From before, mappings are pure functions, but the other two types are not.
Pure functions are analogous to mathematical functions.
]
.pull-right[
.panelset[
.panel[.panel-name[pure functions]
``` r
function(x, y) {
x + y
}
function(y) {
function(x) {
x + y
}
}
```
]
.panel[.panel-name[non-pure functions]
``` r
rnorm(10)
read_csv("file.csv")
function(x) {
Sys.Date() + x
}
function(x) {
x + y
}
function(x) {
function() {
x <<- x + 1
x
}
}
```
]
]
]
---
## Mathematical Functions
.pull-left[
In contrast to the definition of a function in programming, the definition of a function in mathematics is concrete. A
function
`$$f : A \rightarrow B$$`
is a relationship between two set's, `\(A\)` and `\(B\)`, such that every
element from `\(A\)` is mapped to exactly one element in `\(B\)`.
.center[.image-50[

]]
]
.pull-right[
Given two functions,
`$$f: A \rightarrow B\qquad\textrm{and}\qquad g: B \rightarrow C$$`
we can create a new function
`$$H: A \rightarrow C$$`
By composing `\(f\)` and `\(g\)`, we write this as `\(g \cdot f\)` (g after f).
Composition is a powerful tool as it allows us to build complexity by chaining together (simpler) functions.
]
---
class: middle, center
# Why do we care about pure functions?
---
## Referential Transparency
.pull-left[
Pure functions are useful as they can make code significantly easier to debug and test. Pure functions have the property
of [referential transparency][ref_transp], which put simply means we can replace an expression with it's corresponding
value.
For example, consider the function `fn` (shown to the right). Because the function `fn` is pure, we know that it will
always return the same result given the same input, so replacing it's call with the return value is going to yield the
same result.
This can make debugging/testing `another_fn` simpler. The behaviour of `another_fn` is not dependent on `fn`.
]
.pull-right[
``` r
fn <- function(x) {
x + 3
}
```
``` r
another_fn(fn(3), 2)
```
``` r
# we can replace the call to fn() with
# it's return value
another_fn(6, 2)
```
]
---
## Testing
.pull-left[
Knowing that a function will always return the same value given the same inputs makes writing unit tests for the
function significantly easier because:
* We don't need to set up the global environment correctly before running the function - functions that rely on global
state would need to test the function multiple times with the global environment set up with all the possible values.
* We don't need to check the side effects of the function, we just need to check the return value is as expected.
Functions that rely on a side effect can suffer transient errors, e.g. you try to read data from a database, but the
server is temporarily down/busy.
* All we need to do is check that the outputs are correct for given inputs.
]
.pull-right[
``` r
library(testthat)
triangle_number <- function(x) {
0.5 * x * (x + 1)
}
test_that("it works as expected", {
expected_that(triangle_number(1), 1)
expected_that(triangle_number(2), 3)
expected_that(triangle_number(3), 6)
expected_that(triangle_number(4), 10)
expected_that(triangle_number(5), 15)
})
```
]
---
## Parallel Computation
.pull-left[
Writing parallisable code which relies on either shared state or side effect's is notoriously difficult.
Consider the following:
``` r
counter <- 0
increment <- function() {
counter <<- counter + 1
}
```
If we tried to run this function twice in parallel, if both workers try to start at exactly the same time they will both
see `counter` as 0. So both will try to set `counter` to 1, not 2 as may be expected.
That is, we depend on the order of evaluation, and the timing of when they functions are called.
]
.pull-right[
Pure functions however are easy to parallise, because if a function only depends on the arguments that it is provided
then this issue goes away.
The function calls never interact with each other, and can be evaluated in any order, we will always get the same
results as if we evaluated the calls one after another (up to order of results).
We will see later how the `{furrr}` package can help us to parallelise code.
]
---
# Higher order functions
.pull-left[
A higher order function is a function which either:
* takes a function as an argument,
* or, returns a function.
Functions in R are what we call "first class citizens": they are like any other value (such as a numeric vector, or a
character).
As such, we can simply pass a function as an argument just by using it's name, or we can return a function
by creating a function and using that as the return value.
For example, the following is valid code in R:
``` r
my_function <- function(fn, x) {
force(x)
return(function(y) fn(x, y))
}
```
]
.pull-right[
The example given to the left takes a function `fn` which expects two arguments, `x` and `y`, and returns a new function
which always uses the same value for the `x` argument.
This is called **partial application** and can be a very powerful tool in functional programming - we are making a new
function from existing functions.
The `{purrr}` package has a function just for this purpose:
```r
add_three <- purrr::partial(`+`, 3)
add_three(2)
```
```
## [1] 5
```
]
---
## Composition
.pull-left[
If we have 2 (or more) **pure** functions, and we know that the return type for one function is the input type for the next,
then we can build a new function that composes these functions together. `{purrr}` has a function that does this for us,
called `compose`.
``` r
f <- function(x) x * 2
g <- function(x) x + 3
4 %>% f() %>% g() # 11
5 %>% f() %>% g() # 13
h <- compose(g, f)
h(4) # 11
h(5) # 13
```
]
.pull-right[
The one downside to compose is if our functions accept multiple arguments.
To get around this we can combine `compose` and `partial`:
``` r
f <- function(x, y) x * y
g <- function(x, y) x + y
4 %>% f(2) %>% g(3) # 11
5 %>% f(3) %>% g(3) # 18
h <- compose(partial(g, y = 3), f)
h(4, 2) # 11
h(5, 3) # 18
```
]
In some respects, `compose` is like `%>%`: the difference is `compose` creates a new function which can be reused in
other parts of our code easily.
---
# Iteration with loops
.pull-left[
Previously we looked at an example where we used a for loop to run the same function on every column in a dataframe.
We had to iterate in this way because the function that we had (`rescale_01`) worked on individual numeric vectors.
We needed to set up the for loop by first extracting the list of columns, then iterating over each column running the
function.
This is a very common pattern: take a list we want to iterate over, then evaluate some function using that list.
``` r
for(i in colnames(df)) {
df[[i]] <- rescale_01(df[[i]])
}
```
]
.pull-right[
However, it's very easy to make a mistake creating a loop this way: what if we occidentally update the wrong item in the
dataframe?
``` r
for(i in colnames(df)) {
df[[1]] <- rescale_01(df[[i]])
}
```
Or we don't correctly initialise the iteration?
``` r
for(i in 1:4) {
# does our data always have 4 columns?
df[[i]] <- rescale_01(df[[i]])
}
```
]
---
# `purrr::map()`
.pull-left[
The `map` function from `{purrr}` takes a vector/list, and a function. It the evaluates the function once for each
input, returning the results as a list.
We could replace our loop example quite simply with a map function:
``` r
df <- map_dfc(df, rescale_01)
```
The map function's arguments are
``` r
map(x, fn, ...)
```
where
* `x` is the vector you wish to iterate over
* `fn` if the function
* ... are any extra arguments the function requires (these are the same for all calls of the function)
]
.pull-right[
The image, courtesy of [adv-r] shows graphically how the map function works.
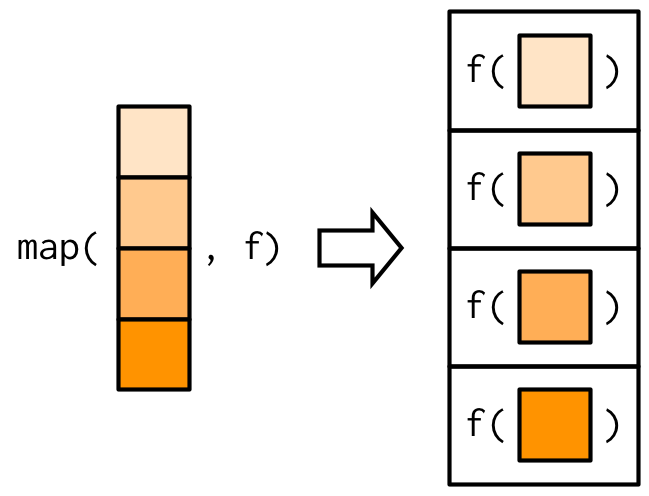
]
---
# `map` in action
.pull-left[
A toy example, let's take a vector of numbers and double them
``` r
values <- 1:5
# we can use a "named function"
double_num <- function(x) 2 * x
map(values, double_num)
# or, we can use an anonymous function
map(values, function(x) 2 * x)
# with R > 4.1, we can use \(x) 2 * x
map(values, \(x) 2 * x)
# we can also use a formula
map(values, ~ .x * 2)
```
]
.pull-right[
Any one of these would return the same thing: a list containing the results
```
## [[1]]
## [1] 2
##
## [[2]]
## [1] 4
##
## [[3]]
## [1] 6
##
## [[4]]
## [1] 8
##
## [[5]]
## [1] 10
```
]
---
# `map_*` variants
.pull-left[
In the previous example we can see the output of `map` is a list. It would be more useful for that function to return a
numeric vector instead. Fortunately, there is a simple way to achieve this in purrr:
```r
map_dbl(values, \(x) 2 * x)
```
```
## [1] 2 4 6 8 10
```
The variants provided are:
* `map_chr` for a function which returns a character
* `map_int`/`map_dbl` for a function which returns an integer/double
* `map_lgl` for a function which returns a logical (`TRUE`/`FALSE`)
* `map_raw` for a function which returns a raw
* `map_df`/`map_dfr`/`map_dfc` for a function which returns a dataframe
]
.pull-right[
There work so long as the function returns a single one of these values so
``` r
length(x) == length(map(x, fn))
```
Now, the example given so far isn't particularly useful: many operations in R are vectorised, so we could just do
`2 * values` and we would get exactly the same results as `map_dbl(values, \(x) 2 * x)`.
`map` functions are useful for cases where we have functions which aren't vectorised and we need to run the function
once for each item in the input vector.
]
---
# `map_df` variants
The data frame variants all do the same sort of thing: if a function returns a dataframe, rather than returning a list
of dataframes, it will bind the results together into a single dataframe:
* `map_df` and `map_dfr` use `bind_rows` to "union" the results together
* `map_dfc` uses `bind_cols` instead
One of the best use cases for `map_df` is to read in a folder full of csv's and combine the results together.
First, let's get a list of all the files in a folder. In this particular folder our files are named `YYYY-MM-DD.csv`.
The dir command with `full.names = TRUE` will return the full file path, but it will be useful to name each item just
after the date part. We use the `set_names()` function from `{purrr}` to achieve this, it accepts a list of items, and
then either a vector of names, or a function to transform the vector by.
```r
files <- dir("data/ae_attendances/", "*.csv", full.names = TRUE) %>%
set_names(function(.x) stringr::str_extract(.x, "\\d{4}-\\d{2}-\\d{2}"))
files[1:2]
```
```
## 2016-04-01 2016-05-01
## "data/ae_attendances/2016-04-01.csv" "data/ae_attendances/2016-05-01.csv"
```
---
## `map_df` variants (continued)
Now that we have our list of files to load, we can use the `read_csv` function to load each csv file. Here, we pass to
the `read_csv` function the column types used in the files, and we also set the `.id` argument of `map_dfr`. This will
create a column in the final dataframe containing the "name" of each item in the list, e.g. the date from the filename.
```r
ae_attendances <- map_dfr(files, read_csv, col_types = "ccddd", .id = "period")
head(ae_attendances, 8)
```
```
## # A tibble: 8 x 6
## period org_code type attendances breaches admissions
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 2016-04-01 RF4 1 18788 4082 4074
## 2 2016-04-01 RF4 2 561 5 0
## 3 2016-04-01 RF4 other 2685 17 0
## 4 2016-04-01 R1H 1 27396 5099 6424
## 5 2016-04-01 R1H 2 700 5 0
## 6 2016-04-01 R1H other 10317 143 0
## 7 2016-04-01 AD913 other 3836 1 0
## 8 2016-04-01 RYX other 17369 0 0
```
---
## Functional Programming in dplyr
.pull-left[
You may have noticed that the `period` column before was a character - it would be much more useful to convert this to a
date.
We could just write a mutate statement like
``` r
ae_attendances %>%
mutate(period = as.Date(period))
```
But, there is a much neater way of writing this out: we can use the `across` function from dplyr to apply a function to
a column.
`across` takes a column specification (either a name of a column, or a function like `where(is.numeric)`, and then a
function to apply to the column(s).
]
.pull-right[
```r
ae_attendances <- ae_attendances %>%
mutate(across(period, as.Date))
head(ae_attendances, 4)
```
```
## # A tibble: 4 x 6
## period org_code type attendances breaches admissions
## <date> <chr> <chr> <dbl> <dbl> <dbl>
## 1 2016-04-01 RF4 1 18788 4082 4074
## 2 2016-04-01 RF4 2 561 5 0
## 3 2016-04-01 RF4 other 2685 17 0
## 4 2016-04-01 R1H 1 27396 5099 6424
```
]
---
# `map2` and `imap`
So far we have looked at cases where we have a single vector to iterate over, but what if we have two vectors, both of
the same length? We can use the `map2` family of functions! There is `map2` which is equivalent to `map`, and then all
of the same `map2_*` variants as we just saw for `map`.
```r
letters <- c("a", "b", "c")
times <- 1:3
map2_chr(letters, times, \(x, y) paste(rep(x, y), collapse = ""))
```
```
## [1] "a" "bb" "ccc"
```
related to `map2` is `imap`. This only accepts a single vector, like `map`, but it creates a second "index" argument. If
the vector is named, this "index" will be the name of the item, otherwise it will be the numerical position.
```r
imap_chr(letters, \(x, y) paste(rep(x, y), collapse = ""))
```
```
## [1] "a" "bb" "ccc"
```
---
# `pmap`
.pull-left[
While `map` works with a single vector, and `map2` works with 2 vectors, `pmap` is a generalised version that works on
any number of vectors. `pmap` also has all of the variants that we have seen before.
Below is a toy example showing how `pmap` works. First, we need to construct a list that contains all of the vectors.
Note, the vectors must all be the same length: we are in effect going to loop over the first item from each vector, then
the second, etc. We then create a function which has one argument per vector.
```r
values <- list(1:3, 4:6, 7:9)
pmap_dbl(values, \(x, y, z) x * y + z)
```
```
## [1] 11 18 27
```
]
.pull-right[
If we instead use a named vector, then it will match the named items in the list to the arguments in the function, for
example, see how we get very different results when wee name the items differently from the order that the arguments
appear in the function.
```r
values <- list(z = 1:3, y = 4:6, x = 7:9)
pmap_dbl(values, \(x, y, z) x * y + z)
```
```
## [1] 29 42 57
```
]
---
# `pmap` in action: individual plots
.pull-left[
Facetted plots in ggplot are great, but what if you ever want to create individual plots and save the files?
First, let's use the `ae_attendances` dataset from the `{NHSRdatasets}` package. This contains 36 months of data of
A&E performance figures, split by trust and department type.
Let's say that we want to create a plot of attendaces for each of the different department types, summarised for every
trust.
Let's create a tibble that contains one row per department type, with a column that contains a nested dataset of all of
the rows of data for that group.
]
.pull-right[
```r
library(NHSRdatasets)
ae_types <- ae_attendances %>%