This repo contains the R statistical code to reproduce the analysis in the paper The health system response to COVID-19 will continue long after we vaccinate 80% of eligible adults (add link to final publication).
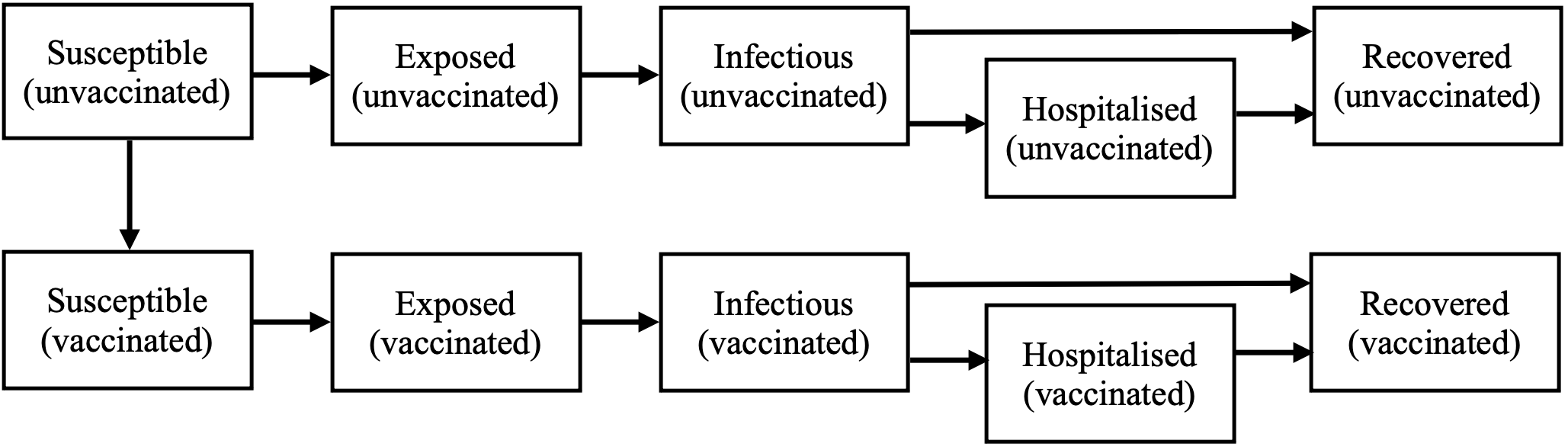
This analysis uses an age-structured deterministic compartmental model implemented in the R package COVOID (COVID-19 Opensource Infectious Dynamics). In this extension of the widely used SIR epidemic model, individual disease status is classified as one of five states: Susceptible, Exposed, Infectious, Hospitalised or Recovered (SEIHR). The model also distinguishes between vaccinated and unvaccinated individuals, resulting in a total of 10 compartments, as depicted below.
Technical details, installation instructions and worked examples for the COVOID R package are available from the companion website https://cbdrh.github.io/covoidance/. A complete description of the model specifcation and assumptions used in this analysis is provided in the Technical Appendix to the main paper.
The analysis was undertaken using R v4.0.3 running on unix (Darwin 17.0) and the following R packages:
-
covoid(0.2.0) -
tidyverse(1.3.1) -
kableExtra(1.3.4) -
ggpubr(0.4.0) -
RColorBrewer(1.1-2)
Hanly MJ, Churches, T, Fitzgerald O, Post JJ, MacIntyre CR, Jorm L. The health system response to COVID-19 will continue long after we vaccinate 80% of eligible adults, The Medical Journal of Australia 2021. (TBC following publication)
All files to replicate, adapt or extend this analysis are saved in the R/ subfolder. There are six files in total:
simulation-control.Rsimulation-functions.Rseir-cv-baseline.Rseir-cv-scenarios-noNPI.Rseir-cv-scenarios-NPI.Rseir-cv-visualisations.R
The entire analysis can be reproduced by running the file simulation-control.R. This script will call the other files in the order listed below to run the simulations from the published analysis and recreate the corresponding figures. Adapting or extending the analysis requires editing the other files as desired. The purpose of each script is brielfy summarised below.
simulation-control.R
- Loads required libraries.
- Sets a random seed.
- Creates an
Outputssubfolder, if it does not exist. - Sources the remaining files in order to replicate the published analysis.
simulation-functions.R
- Defines useful functions used in the analysis and data summary.
seir-cv-baseline.R
- Defines a baseline parameterisation of the SEIHR epidemic model.
seir-cv-scenarios-noNPI.R
-
Parameterises and runs the simulations for scenarios without non-pharmaceutical interventions (NPIs) on social contacts.
-
Saves the following files in the
Outputsfolder:simulation_results_noNPI.Rda(A list with the raw results of each simulation run)incidence_results_noNPI.Rda(A list with a condensed summary of each simulation run)incidence_results_full_noNPI.Rda(The condensed summary of all simulations compiled into a dataframe)
seir-cv-scenarios-NPI.R
-
Parameterises and runs the simulations for scenarios with nNPIs on social contacts.
-
Saves the following files in the
Outputsfolder:simulation_results_NPI.Rda(A list with the raw results of each simulation run)incidence_results_NPI.Rda(A list with a condensed summary of each simulation run)incidence_results_full_NPI.Rda(The condensed summary of all simulations compiled into a dataframe)
seir-cv-visualisations.R
-
Creates plots of the vaccine rollout projections, and estimates hospitalisation & infection over time.
-
Specifically, saves the following files in the output folder:
yyyy-mm-dd-vaccine_projections.png(projected vaccination coverage over time)yyyy-mm-dd-hospitalisation_projections.png(projected number of hospitalised patients over time)yyyy-mm-dd-hospitalisation_projections-hi-res.png(projected number of hospitalised patients over time ,higher resolution)yyyy-mm-dd-infection_projections.png(projected number of infected patients over time)yyyy-mm-dd-infection_projections-hi-res.png(projected number of infected patients over time, higher resolution)
An early version of this analysis was published as an MJA preprint at the following link: https://www.mja.com.au/journal/2021/health-system-response-covid-19-will-continue-long-after-we-reach-80-vaccination