Integrating Medical AI Compute CPU/GPU workflows on the MOC -- PowerPC and x86-64 -- Using OpenShift
Project Members: Elaine Parr, Mohit Chandarana, Reed Hansen, Ziyu Liu
With the fast development of computing technologies in recent decades, there has been a huge amount of work in the medical image research area. However, these works are rarely used in practice at scale, due to the heterogeneity of deployment used by different researchers. ChRIS (Children’s Research Integration Service) developed at Boston Children's Hospital is a general-purpose compute/data platform that simplifies deploying analysis on a heterogeneous mix of computer environments including workstations, local networks, HPC clusters, and public clouds. Our purpose is to enable ChRIS to utilize the rich computing resources of the Massachusetts Open Cloud (MOC).
The primary goal of this project is to enable the execution of existing ChRIS workflows on the MOC, specifically on the x86-64 cluster. The ChRIS Ultron Backend (CUBE) interacts with a compute service and manages compute tasks such as neuroimaging analysis, COVID detection from CT scans, etc. Our goal is to build production level ancillary services on the MOC that allow the CUBE to run compute tasks on MOC. The process and controller utilities (pman and pfcon) run in a Docker swarm environment locally. When running on the MOC, these services need to run in an OpenShift environment; both the environment and the services need to be properly configured for these services to be deployed and run.
Additional goals are included in the acceptance criteria section.
The users of our project will be ChRIS developers and other power users.
ChRISDevelopers will use the results of our project to provide the option of executing tasks on the MOC for new workflows, as well as additional functions around using the MOC- Power users like admins will use the results of our project to incorporate new algorithms (packaged as plugins) that utilize the compute resources of the MOC
Note: the ChRIS service does not directly target downstream users like clinicians and life science researchers. Services available to them will have specific use cases (such as the COVID-Net workflow) and these services will call ChRIS to do computation in the backend.
The scope of this project is generally adapting existing features to function on the MOC. Adapting the COVID-Net workflow to run on the MOC will be the primary goal. We hope to add a few novel features, such as allowing users to choose between computing in the local environment or in the MOC. Additional goals include enhancing COVID-Net workflow with new plugins. Features to create:
- Adapt the existing workflow management functionally of the
pmanmodule toOpenShift - Allow for each task in a workflow to be run on in a local Docker swarm or in
OpenShift - Develop new plugins and add them to the existing workflow
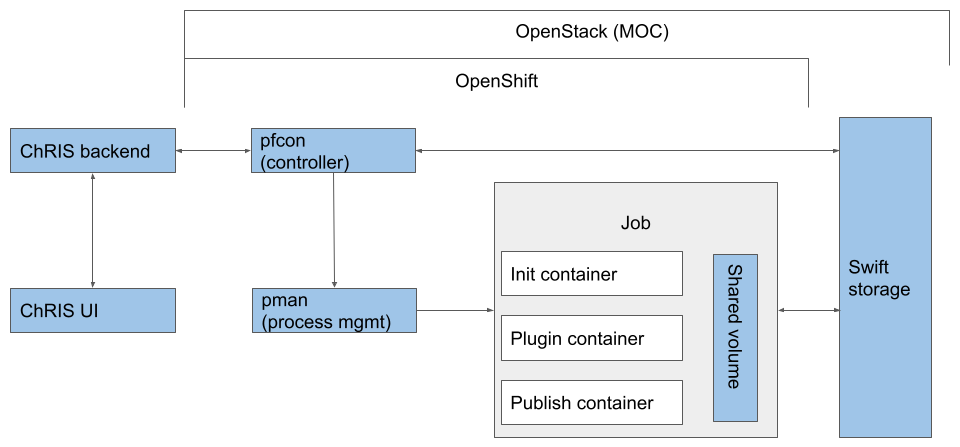
Below is a diagram of the system components with MOC incorporated.
The ChRIS UI and backend are hosted locally or within the institutional network. Two ChRIS components, pfcon and pman, are hosted on MOC OpenShift. Compute tasks are actually executed on MOC OpenShift as well.
In production, an application like COVID-Net is packaged as a tree of “plugins”. When a plugin run is requested from ChRIS, CUBE sends the request to remote ancillary services, which handle pulling the image to run, scheduling compute and data transfer.
Note: The service structure described below is the new version that was rolled out during the semester. Our major contribution is enabling the deployment of these rewritten services on OpenShift and enabling running plugins with CUBE talking to these deployed instances.
Two services need to be deployed in OpenShift such that CUBE can run tasks with them.
The controller, pfcon, is a controlling service that acts as the interface to a remote pman service. In the data plane, it handles file upload/download and zipping/unzipping. In the control plane, it schedules a plugin run with pman, checks on the run status, and sends the status back to ChRIS.
The process manager, pman, provides a unified API over HTTP for running jobs on Docker Swarm and Kubernetes/OpenShift. Upon request, pman schedules k8s job pods. In each job pod, there are three containers that run sequentially and share a volume: the init container pulls data from Swift, the job container runs the plugin on the data, and the publish container pushes data back to Swift.
This structure also enables crossover runs. After a plugin run completes, the result data is transferred back to where ChRIS is. When multiple pfcons are registered on ChRIS, the user can specify which environment a plugin is run; ChRIS then sends the specifications and data to the said environment. So in a crossover run, the user can run a plugin on a local environment, then run a next plugin (which may need better computing capacities or specialized hardware) on the MOC. Such crossover runs grant users great flexibility in scheduling compute tasks, therefore running plugins are easier and more efficient.
The structure presented above is version 3. During the semester, the ChRIS team at BCH redesigned the service structure and rewrote the ancillary services. In the old structure (version 2), file operations are handled in a separate service called pfioh. In the new structure, pfcon took this responsibility so pfioh is no longer in the structure. Also, all services are rewritten in flask, but none of the config files or deployment templates are updated. We worked with the ChRIS team to enable deploying the new service images onto OpenShift, and we are working to deploy them on MOC OpenShift.
ChRIS offers containerized computations called plugins. These are standalone programs that can run with or without ChRIS (using docker). Plugins may perform data transformations or analyses (computes) that can be chained together into a workflow.
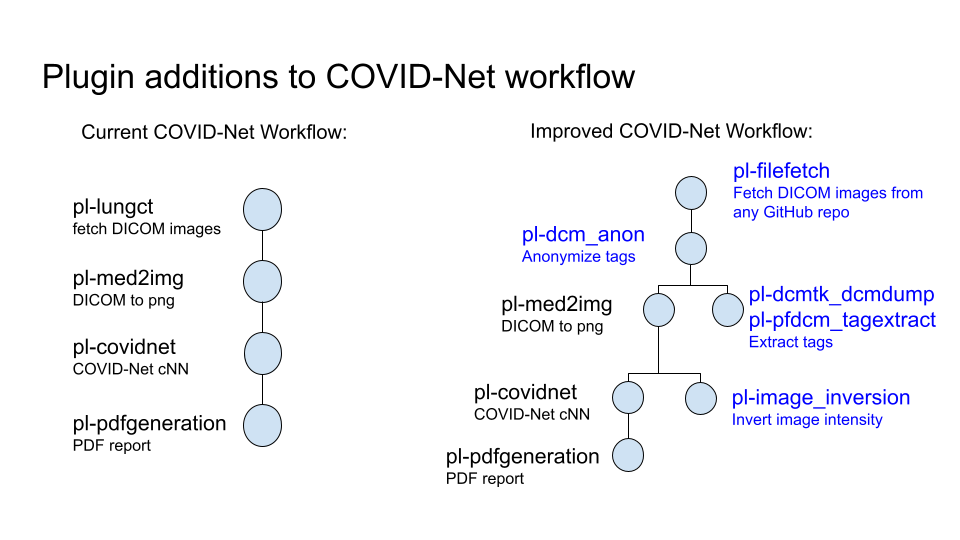
The focus of this project is the COVID-Net workflow, which analyses lung scans using a Neural net to predict whether a given patient has COVID-19. There are 4 plugins in the current COVID-Net workflow -
pl-lungct -> pl-med2img -> pl-covidnet -> pl-pdfgeneration
- pl-lungct - fetches a set of lung images (in DICOM format)
- pl-med2img - converts a DICOM image to png
- pl-covidnet - runs the png files through a neural network and generates predictions
- pl-pdfgeneration - takes the predictions and generates a pdf report
As part of our stretch goals, we decided to add a few plugins to the COVID-Net workflow. The figure below shows the proposed changes to the current workflow:
Plugin additions to the COVID-Net workflow:
- pl-lungct2
- provides additional DICOM images for the COVID-Net workflow
- pl-filefetch
- fetches files of a specified type from a list of GitHub repositories or a subdirectory of repositories
- pl-dcm_anon
- takes input directory of DICOM images and anonymizes sensitive/identifying patient information in the tags
- pl-dcmdump
- extracts metadata (tags) from DICOM images using the
dcmdumputility offered bydcmtk(DICOM toolkit) into a text file
- extracts metadata (tags) from DICOM images using the
- pl-pfdcm_tagextract
- extracts metadata (tags) from DICOM images
- The output can be stored in the following file formats:
- raw, json, html, dict, col, csv
- pl-image_inversion
- inverts the colors of an image and can convert the output to a different image format.
The minimum acceptance criteria is to enable the execution of the COVID-Net workflow using the ChRIS UI or scripts. The backend services should run on OpenShift.
Stretch goals include:
- Get all of the workflows to run on the MOC using
OpenShiftin x86-64 - Enable crossover run: running any workflow node locally or on
OpenShift, with data seamlessly transferred from one node to another - Create anonymization plugin
- Create new lungct plugin
- Create filefetch plugin
- Create image inversion plugin
- Create tag extraction plugins
This section lists our contribution to the ChRIS project, as well as our demo.
- https://github.com/FNNDSC/ChRIS-E2E (pfcon test scripts)
- https://github.com/FNNDSC/pman (pman deployment template and config update)
- https://github.com/FNNDSC/pl-covidnet (covidnet plugin fix)
- https://github.com/LilMit/pl-dcm_anon (anonymization plugin git repo)
- https://hub.docker.com/repository/docker/parrmi/pl-dcm_anon (anonymization plugin dockerhub)
- https://github.com/mohitchandarana/pl-dcmtk_dcmdump (pl-dcmtk_dcmdump git repo)
- https://hub.docker.com/repository/docker/mchandarana/pl-dcmtk_dcmdump (pl-dcmtk_dcmdump dockerhub)
- https://github.com/mohitchandarana/pl-pfdcm_tagextract (pl-pfdcm_tagextract git repo)
- https://hub.docker.com/repository/docker/mchandarana/pl-pfdcm_tagextract (pl-pfdcm_tagextract dockerhub)
- https://hub.docker.com/repository/docker/reedh13/pl-lungct2 (pl-lungct2 dockerhub)
- https://github.com/reedh13/pl-lungct2 (pl-lungct2 git repo)
- https://github.com/reedh13/pl-image_inversion (pl-image_inversion git repo)
- https://hub.docker.com/repository/docker/reedh13/pl-image_inversion (pl-image_inversion dockerhub)
- https://hub.docker.com/repository/docker/reedh13/pl-filefetch (pl-filefetch dockerhub)
- https://github.com/reedh13/pl-filefetch (pl-filefetch git repo)
- https://github.com/LilMit/pl-dcm_anon (anonymization git repo)
- https://hub.docker.com/repository/docker/parrmi/pl-dcm_anon (anonymization dockerhub)
- https://github.com/LilMit/CHRIS_docs/blob/master/workflows/covidplugins_upload.sh (helper script to add COVID_net workflow plugins to ChRIS)
- https://github.com/LilMit/CHRIS_docs/blob/master/workflows/covidnet_anonymized.sh new covidnet shell script with anonymization plugin
- https://github.com/FNNDSC/ChRIS_on_MOC/wiki (instructions on deploying ChRIS services on MOC)
- https://github.com/Cagriyoruk/CHRIS_docs/blob/master/usecases/MOC_integration/moc_integration.adoc (more instructions including local OpenShift instantiation)
- FNNDSC/CHRIS_docs#12 (instructions for ChRIS and COVID-Net on WSL)
- FNNDSC/pl-med2img#2 (updates to old docs for med2img)
- FNNDSC/python-chrisclient#3 (updates to README for python-chrisclient)
https://drive.google.com/file/d/1J3EPFdr8XdnXrsGZff5hrNSo8bKMLNbw/view?usp=sharing
- Sprint 1:
- ChRIS backend local installation
- Familiarizing ChRIS structure
- Learning OpenShift
- Sprint 2:
- Pack deployment-ready pman and pfioh images
- Deploy these images on MOC OpenShift
- Sprint 3:
- New version of pfcon/pman out, pack, deploy and test them
- Start working on plugins - MOC outage
- Sprint 4:
- Run COVID-Net workflow on local OpenShift cluster
- Run COVID-Net workflow through full ChRIS - pfcon - pman stack
- Finish developing plugins and test them locally
- Sprint 5:
- Add new plugins to COVID-Net workflow