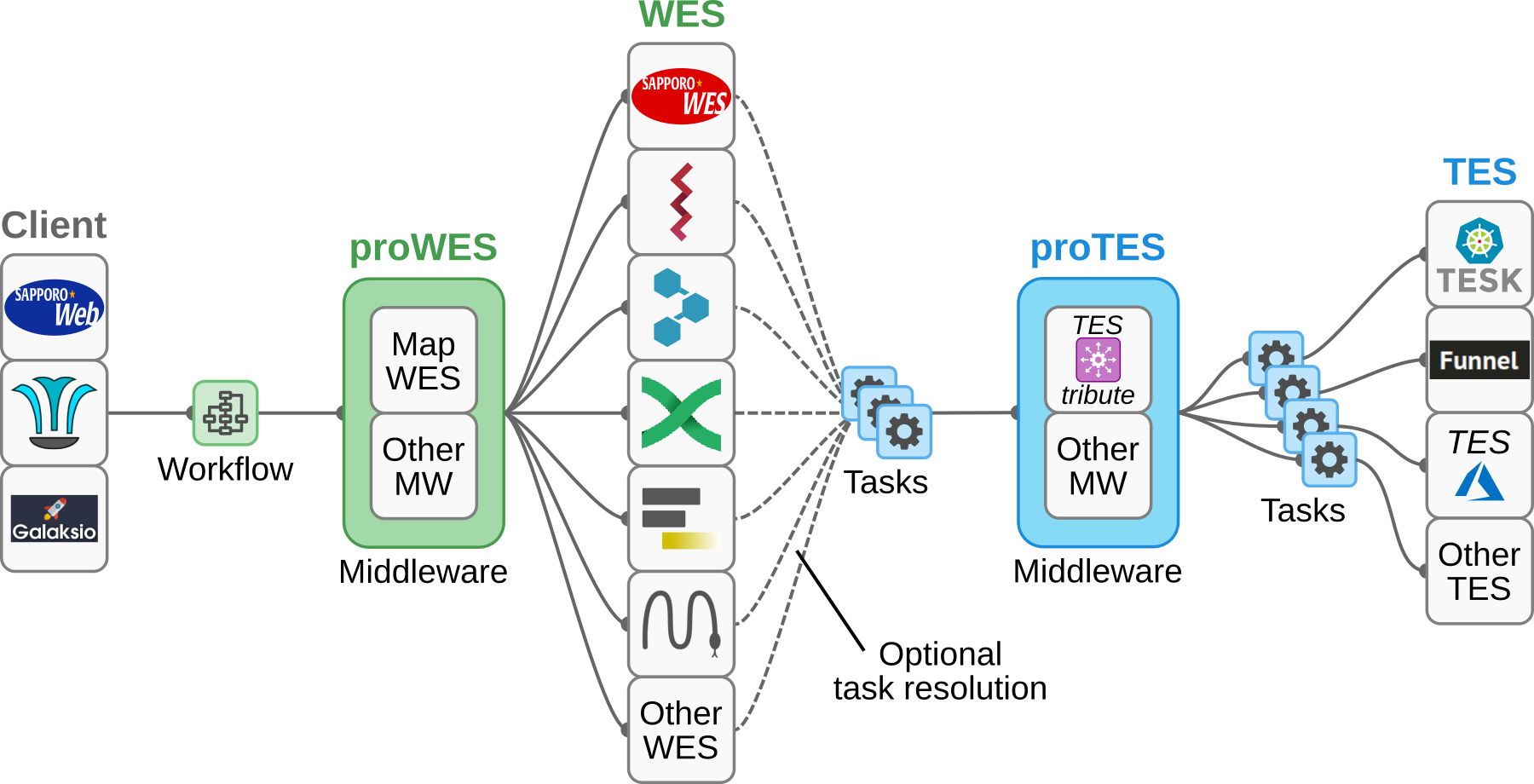
This is a proof of concept to invoke a galaxy workflow in a galaxy container using the WES API. This will act as the proWES part (middleware) in the picture.
A Galaxy 19.01 container is decorated with the necessary tools to run the workflow. This galaxy container contains both the workflow and task execution service.
docker build -t containername -f Dockerfile .Only workflows that are pre installed (at this moment the ones that are located in the /workflow folder) will be supported. In the future, supported workflows will all have their own image. Supported workflows will be based on an upcoming workflow registry.
To mimic the WES /runs POST call, following example json is used as input:
{
"workflow_params": {
"inputs": [
{
"step": "0",
"file": "https://raw.githubusercontent.com/bedroesb/WES2Galaxy/master/example_data/UCSC_input.bed"
}
]
},
"workflow_url": "https://raw.githubusercontent.com/bedroesb/WES2Galaxy/Dev/example_data/Galaxy-Workflow-galaxy-intro-strands-2.ga",
"workflow_version": "0.1",
"workflow_type": "ga"
}
In the future, this WES call will create a new workflow run and will return a RunId to monitor its progress.
-
The workflow_url is an absolute URL to a workflow file.
-
The workflow_params JSON object specifies input parameters, such as input files. The exact format of the JSON object depends on the conventions of the workflow language being used. Input files should either be absolute URLs.
-
The workflow_type is the type of workflow language.
-
The workflow_type_version is the version of the workflow language submitted and must be one supported by this WES instance.
The attribute inputs contains a list of all the input files with their corresponding file and workflow step. Note that the step number is in string format.
A Galaxy 19.01 container will be decorated with the necessary tools to run the workflow.
To build the docker image use following command in the root directory:
docker build -t containername -f Dockerfile .To run the container interactively use following command:
docker run -t containername- Using a WES flask client to invoke the workflow in the galaxy container
- Implementing the possibility to check the workflow status
- Kill te container after specific amount of time (when user has data)
- Give ID back to the user for retrieving status information
- Separating the workflow execution service in galaxy from the task execution service. (This is already possible)
- How to supply the output-data in the container back to the user?
-> Available on a server for a limited amount of time.
-> Downloadable through links - When the
startingWorkflow.pyscript is manually started in the container, BioBlend is not installed. No problem when the script is started automatically with
ENTRYPOINT [ "/bin/starter-service.sh" ]
in the Dockerfile ? - Not working with latest galaxy docker 19.05