This is an example of how to set up a data model using the Software-Driven Research Data Management (sdRDM) library which is based on abstract object models. Furthermore, the sdRDM library supports the conversion of data models defined in the Markdown format.
Data models defined in the Markdown format follow these conventions:
- Title of the data model is denoted by a heading level 1
# - Modules are denoted by a heading level 2
## - Objects are started with a heading level 3
### - Each object contains fields as a list →
- name - Required fields are set by making the field bolt →
- __name__ - Each field has options as a list of name to value mapping →
- Type: string
Each field in an object can hold options relevant for mapping to another data model (e.g. a standardized format) and general information such as its type and description. In the following is a collection of all native and required fields:
- Type - Required option to denote the data type. Please note, this can also contain other objects defined in this document.
- Multiple - Whether or not this field can contain multiple values. Setting to
Truewill result in aList[dtype]annoatation in the software. - Description - Required option to describe the field. This should be a brief description that explains what the attribute is about.
In order to inherit attributes to another object, the object definition additionally includes the name of the parent object in italic wrapped with brackets:
→ ## Child [_Parent_]
In the following an example data model is defined using above rules. Feel free to use this example also as a template for your own application.
You can experiment and use this example repository right away to get familiar with the concept. This repository includes an action that is triggered whenever changes are pushed. Thus, when you introduce changes to the markdown document, these will directly be reflected onto the generated software. Follow these steps to start out:
- Fork this repository into your own profile. This will create an exact copy, but you have all rights to modify it without affecting the original.
- Open the
specifications/Example.mdfile and edit it according to the syntax. You can also pressPreviewto inspect the rendered Markdown.
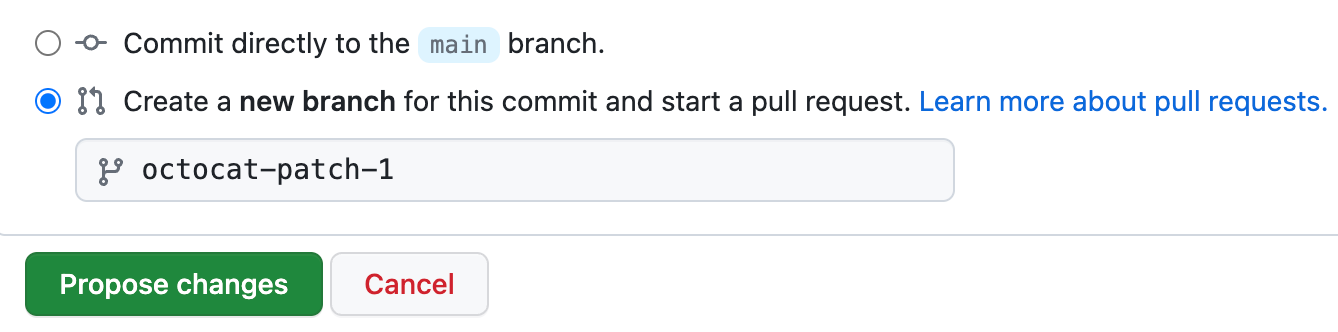
- Commit changes to the
mainbranch or create a new one from it. By creating a new branch you can safely work without affecting the original. Once your modifications are done, you can merge these into themainbranch.
- Watch your changes being reflected onto the API. You can also directly fetch this model using the sdRDM library. For this, you can use the following example code that should run as is.
from sdRDM import DataModel
lib = DataModel.from_git(
url="https://github.com/JR-1991/sdrdm-template.git",
)
# Visualize the data model
lib.Root.meta_tree()
# Enter your data
dataset = lib.Root(title="Some Title", description="Some Description")
dataset.add_to_authors(name="Jan Range", affiliation="SimTech")
dataset.add_to_parameters(key="Param", value=10.0)
# Inspect your dataset
print(dataset.yaml())
# Option: Link your dataset to an option --> Dataverse
dataset.to_dataverse()
# Option: Export your dataset to another format
with open("my_dataset.json", "w") as f:
f.write(dataset.json())
# Re-opening your dataset using sdRDM will cause the library
# to re-build the software state in which the dataset was created(Images were taken from GitHub's "Editing Files" tutorial)