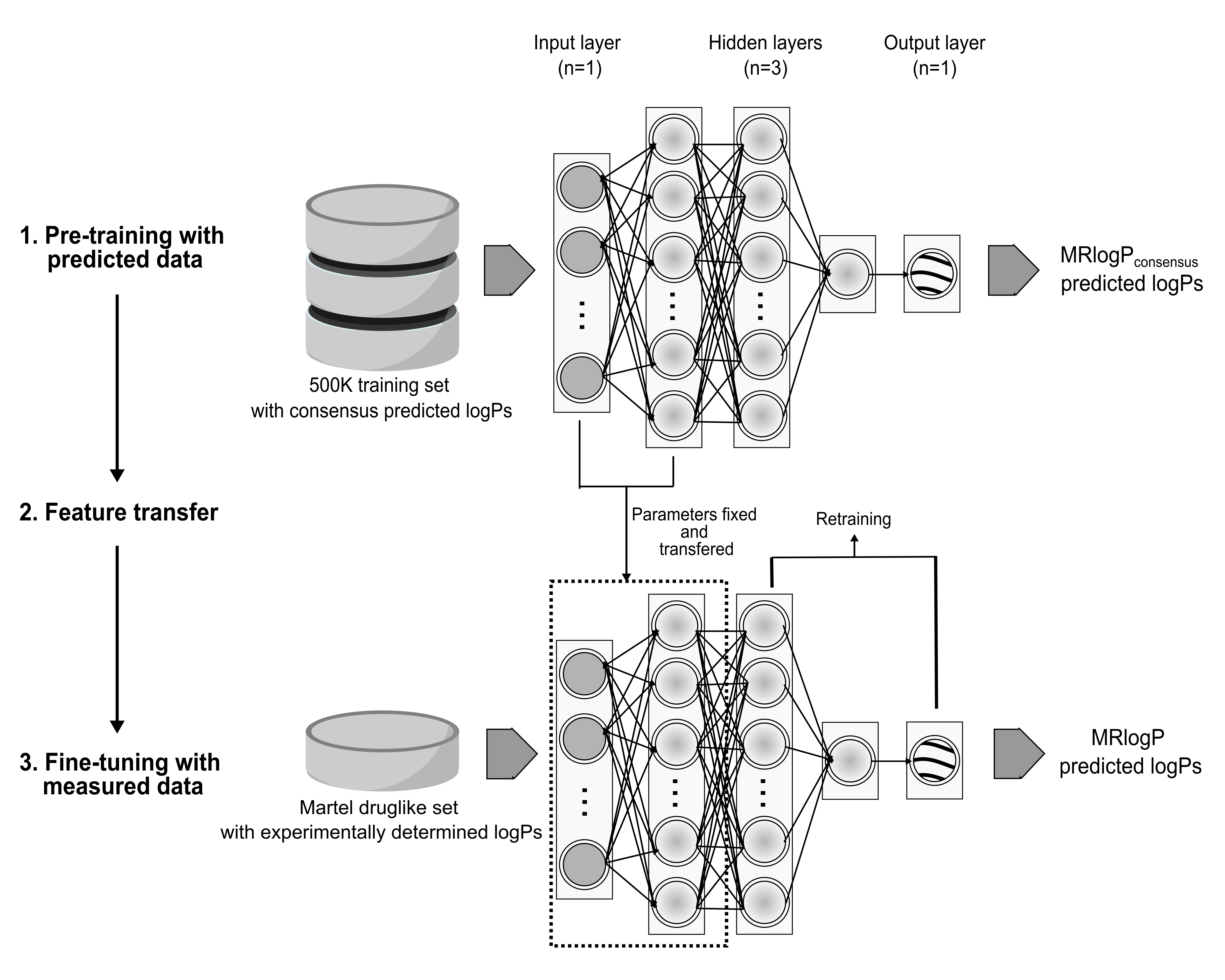
Neural network-based logP prediction for druglike small molecules.
MRlogP is a neural network-based predictor of small molecule lipophilicity (logP), created using transfer learning techniques, demonstrating accurate physicochemical property prediction through training on a large amount of low accuracy, predicted logP values before final tuning using a small accurate dataset of 244 druglike molecules. MRlogP is capable of outperforming state of the art freely available logP prediction methods for druglike small molecules, achieving an average RMSE of 0.988 and 0.715 against druglike molecules from Reaxys and PHYSPROP datasets respectively.
All associated code for MRlogP is freely available to run locally, and additionally available as a simple and easy to use online web-based tool at https://similaritylab.bio.ed.ac.uk/mrlogp.
MRlogP requires the following packages:
- rdkit (>=2019.09.x.x)
- OpenBabel (>=2.4.x)
- numpy (>=1.19.x)
- pandas (>=0.24.x)
- scikit-learn (>=0.20.x)
- TensorFlow (>=2.2.x)
- Keras (>=2.2.x)
Before using MRlogP, please make sure the training set cloned from the repo is uncompressed and merged into a complete csv file.
A quickstart example for the prediction of logP on 5 molecules in the example folder using MRlogP is as follows: In terminal:
python train_mrlogp_model.py pred_logP
Other functions are able to be performed using the parameters in train_mrlogp_model.py. Please see the parameters and the sub-parameters of which as well as examples below for further uses:
para_scan(Required) Perform hyperparameter scan with given a set of hyperparameter combinations using gird search.-ld, --large_dataset(Optional, default=./datasets/ds_descriptors_500K_DL.csv) Dataset containing MRlogP descriptors and logPs used for model training.-para_dict, --hyperparameter_dict(Optional, default=./example/hyperparameter_list.txt) A file contains a dictionary includes set hyperparameters used to perform hyperparameter scan.-val_split, --training_test_split(Optional, default=0.1) Fraction for splitting a subset from training set for validation.
cv(Required) Perform cross validation with given a set of hyperparameter combinations.-ld, --large_dataset(Optional, default=./datasets/ds_descriptors_500K_DL.csv) Dataset containing MRlogP descriptors and logPs used for model training.-para_dict, --hyperparameter_dict(Optional, default=./example/hyperparameter_list.txt) A file contains a dictionary includes set hyperparameters used to perform cross validation.-cv, --n_fold_cross_validation(Optional, default=10) Number of folds for cross validation.
f_train(Required) Train the neural network model with the given hyperparameters against the full training set and then test against test sets.-ld, --large_dataset(Optional, default=./datasets/ds_descriptors_500K_DL.csv) Dataset containing MRlogP descriptors and logPs used for model training.-md, --small_precise_dataset(Optional, default=./datasets/ds_descriptors_martel_DL.csv) Small set of MRlogP descriptors containing highly accurate (measured) used to evaluate the model at the end of the training.-rd, --reaxys_dataset(Optional, default=./datasets/ds_descriptors_reaxys_DL.csv) 2nd test set used to evaluate the model at the end of the training.-pd, --physprop_dataset(Optional, default=./datasets/ds_descriptors_physprop_DL.csv) 3rd test set used to evaluate the model at the end of the training.-mod_file, --model_file(Optional, default=./example/models/mrlogp_model.hdf5) Model file of an exsisting model to be evaluated against the test sets.
t_train(Required) Perform transfer learning on a pre-trained model with given a set of transfer learning hyperparameters.-ld, --large_dataset(Optional, default=./datasets/ds_descriptors_500K_DL.csv) Dataset containing MRlogP descriptors and logPs used for model training.-md, --small_precise_dataset(Optional, default=./datasets/ds_descriptors_martel_DL.csv) Dataset containing high quality measured logPs used to futher tweak the pre-trained model for higher prediction performance.-rd, --reaxys_dataset(Optional, default=./datasets/ds_descriptors_reaxys_DL.csv) Test set used to evaluate the model at the end of transfer learning.-pd, --physprop_dataset(Optional, default=./datasets/ds_descriptors_physprop_DL.csv) 2nd test set used to evaluate the model at the end of transfer learning.-mod_file, --model_file(Optional, default=./example/models/mrlogP_model_consensus.hdf5) The path of the pre-trained model needed for transfer learing.
pred_logp(Required) Predict logP on query compounds using given model.-ld, --large_dataset(Optional, default=./datasets/ds_descriptors_500K_DL.csv) Dataset containing MRlogP descriptors and logPs used for model training.-q_mol, --query_file(Optional, default=./example/sample_5cpd.csv) Descriptor file of query compounds for the logP prediction.-mod_file, --model_file(Optional, default=./example/models/mrlogp_model.hdf5) The path of the model used as the predictor performing logP prediction.
Example 1: Hyperparameter scan
#Hyperparameter scan with the default setting
python train_mrlogp_model.py para_scan
#Hyperparameter scan with customised hyperparameter combinations and 20% of training set
python train_mrlogp_model.py para_scan -para_dict ./example/hyperparameter_c1_list.txt -val_split 0.2
Example 2: Cross validation
#Cross validation with the default setting
python train_mrlogp_model.py cv
#5-fold cross validation with customised hyperparameter combinations
python train_mrlogp_model.py cv -para_dict ./example/hyperparameter_c1_list.txt -cv 5
Example 3: Final training
#Final training with the default setting
python train_mrlogp_model.py f_train
Example 4: Transfer learning
#Transfer learning with the default setting
python train_mrlogp_model.py t_train
#Transfer learn on Physprop datasets and evaluated on Martel and Reaxys datasets
python train_mrlogp_model.py t_train -md ./datasets/ds_descriptors_physprop_DL.csv -rd ./datasets/ds_descriptors_martel_DL.csv -pd ./datasets/ds_descriptors_reaxys_DL.csv
Example 5: LogP Prediction
#Predict molecular logP on the SMILES file in the example folder using the mrlogP model
python smi_to_logP_descriptors.py ./example/sample_5cpd_from_SMILES.smi
python train_mrlogp_model.py pred_logp -q_mol ./example/sample_5cpd_from_SMILES.csv
#Predict logP on 1 molecule in the example folder using the mrlogP consensus model
python train_mrlogp_model.py pred_logp -q_mol ./example/sample_1cpd.csv -mod_file ./example/mrlogP_model_consensus.hdf5
Chen, Yan-Kai, Steven Shave, and Manfred Auer. 2021. "MRlogP: Transfer Learning Enables Accurate logP Prediction Using Small Experimental Training Datasets" Processes 9, no. 11: 2029. https://doi.org/10.3390/pr9112029