-
-
Notifications
You must be signed in to change notification settings - Fork 19
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
* api function (lir) * docstring * setup.cfg
- Loading branch information
1 parent
29343d5
commit 16b35bc
Showing
38 changed files
with
402 additions
and
513 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,88 +1,156 @@ | ||
| # lir | ||
| Largest Interior Rectangle implementation in Python. | ||
|
|
||
| <img height="75" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/sample1.png" /> <img height="75" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/sample2.png" /> <img height="75" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/sample3.png" /> <img height="75" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/sample4.png" /> <img height="75" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/sample5.png" /> | ||
| Fast Largest Interior Rectangle calculation within a binary grid. | ||
|
|
||
| :rocket: Through [Numba](https://github.com/numba/numba) the Python code is compiled to machine code for execution at native machine code speed! | ||
|    | ||
|
|
||
| ### Acknowledgements | ||
| :rocket: Through [Numba](https://github.com/numba/numba) the Python code is compiled to machine code for execution at native machine code speed! | ||
|
|
||
| ## Installation | ||
|
|
||
| Use pip to install largestinteriorrectangle from [PyPI](https://pypi.org/project/largestinteriorrectangle/). | ||
|
|
||
| ```bash | ||
| pip install largestinteriorrectangle | ||
| ``` | ||
|
|
||
| ## Usage | ||
|
|
||
| ```python | ||
| import largestinteriorrectangle as lir | ||
| import numpy as np | ||
|
|
||
| grid = np.array([[0, 0, 1, 0, 0, 0, 0, 0, 0], | ||
| [0, 0, 1, 0, 1, 1, 0, 0, 0], | ||
| [0, 0, 1, 1, 1, 1, 1, 0, 0], | ||
| [0, 0, 1, 1, 1, 1, 1, 1, 0], | ||
| [0, 0, 1, 1, 1, 1, 1, 1, 0], | ||
| [0, 1, 1, 1, 1, 1, 1, 0, 0], | ||
| [0, 0, 1, 1, 1, 1, 0, 0, 0], | ||
| [0, 0, 1, 1, 1, 1, 0, 0, 0], | ||
| [1, 1, 1, 1, 1, 1, 0, 0, 0], | ||
| [1, 1, 0, 0, 0, 1, 1, 1, 1], | ||
| [0, 0, 0, 0, 0, 0, 0, 0, 0]], | ||
| "bool") | ||
|
|
||
| lir.lir(grid) # array([2, 2, 4, 7]) | ||
| ``` | ||
|
|
||
| For [significant performance enhancement](#lir-based-on-contour) in larger grids specify the contours(s) of the polygons to consider. | ||
| If the grid only has one polygon like in the example the contour can be obtained as so (with [opencv](https://pypi.org/project/opencv-python/)). | ||
|
|
||
| ```python | ||
| import cv2 as cv | ||
| cv_grid = grid.astype("uint8") * 255 | ||
| contours, _ = \ | ||
| cv.findContours(cv_grid, cv.RETR_TREE, cv.CHAIN_APPROX_NONE) | ||
| contour = contours[0][:, 0, :] | ||
| ``` | ||
|
|
||
| then calculate the rectangle. | ||
|
|
||
| ```python | ||
| lir.lir(grid, contour) # array([2, 2, 4, 7]) | ||
| ``` | ||
|
|
||
| ## Contributing | ||
|
|
||
| Pull requests are welcome. For major changes, please open an issue first to discuss what you would like to change. | ||
|
|
||
| Please make sure to update tests as appropriate. | ||
|
|
||
| Run tests using | ||
|
|
||
| ```bash | ||
| python -m unittest | ||
| ``` | ||
|
|
||
| Build with | ||
|
|
||
| ```bash | ||
| python -m build | ||
| ``` | ||
|
|
||
| ## License | ||
|
|
||
| [Apache License 2.0](https://github.com/lukasalexanderweber/lir/blob/main/LICENSE) | ||
|
|
||
| ## Acknowledgements | ||
|
|
||
| Thanks to [Tim Swan](https://www.linkedin.com/in/tim-swan-14b1b/) for making his Largest Interior Rectangle implementation in C# [open source](https://github.com/Evryway/lir) and did a great [blog post](https://www.evryway.com/largest-interior/) about it. The first part was mainly reimplementing his solution in Python. | ||
|
|
||
| The used Algorithm was described 2019 in [Algorithm for finding the largest inscribed rectangle in polygon](https://journals.ut.ac.ir/article_71280_2a21de484e568a9e396458a5930ca06a.pdf) by [Zahraa Marzeh, Maryam Tahmasbi and Narges Mireh](https://journals.ut.ac.ir/article_71280.html). | ||
|
|
||
| Thanks also to [Mark Setchell](https://stackoverflow.com/users/2836621/mark-setchell) and [joni](https://stackoverflow.com/users/4745529/joni) who greatly helped optimizing the performance using cpython/numba in [this SO querstion](https://stackoverflow.com/questions/69854335/optimize-the-calculation-of-horizontal-and-vertical-adjacency-using-numpy) | ||
|
|
||
| ### How it works | ||
| ## How it works | ||
|
|
||
| For a cell grid: | ||
| For a binary grid: | ||
|
|
||
| <img width="200" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/cells.png"> | ||
|  | ||
|
|
||
| We can specify for each cell how far one can go to the right and to the bottom: | ||
|
|
||
| Horizontal Adjacency | Vertical Adjacency | ||
| :-------------------------:|:-------------------------: | ||
| <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/h_adjacency.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/v_adjacency.png" /> | ||
| 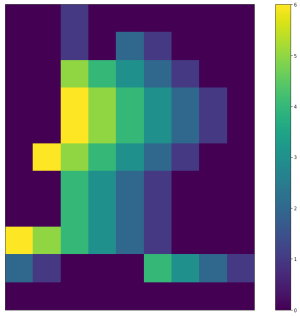 |  | ||
|
|
||
| Now the goal is to find the possible rectangles for each cell. For that, we can specify a Horizontal Vector based on the Horizontal Adjacency and Vertical Vector based on the Vertical Adjacency: | ||
|
|
||
| Horizontal Vector (2,2) | Vertical Vector (2,2) | ||
| :-------------------------:|:-------------------------: | ||
| <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/h_vector.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/v_vector.png" /> | ||
|  |  | ||
|
|
||
| So at the given cell (2,2) the Horizontal Vector is (5,4) and the Vertical Vector is (7,4). | ||
|
|
||
| Reversing either vector lets you create the spans by stacking the vectors, so for example reversing the Vertical Vector to (4,7) gives a set of spans of (5 by 4) and (4 by 7). | ||
|
|
||
| Since `4*7=28 > 5*4=20` a rectangle with width 4 and height 7 is the biggest possible rectangle for cell (2,2). | ||
| The width and height is stored in a span map, where the widths and heights of the maximum rectangles are stored for all cells. | ||
| Using the area we can identify the biggest rectangle at (2, 2) with width 4 and height 7. | ||
| Using the area we can identify the biggest rectangle at (2, 2) with width 4 and height 7. | ||
|
|
||
| Widths | Heights | Areas | ||
| :-------------------------:|:-------------------------:|:-------------------------: | ||
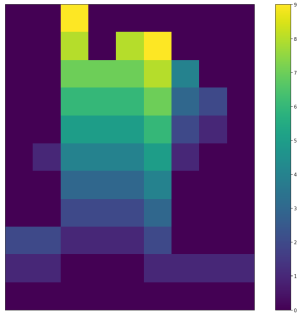
| <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/span_map_widths.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/span_map_heights.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/span_map_areas.png" /> | ||
|  | 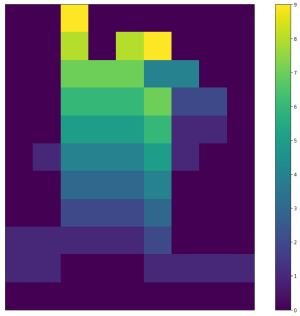 |  | ||
|
|
||
| ------------ | ||
|
|
||
| ### LIR based on outline | ||
| ## LIR based on contour | ||
|
|
||
| Especially for bigger grids the functionality can be further optimized by only analysing the outline. Here are timings created by calculating the lir for [masks in different resolutions](https://github.com/lukasalexanderweber/lir/tree/main/ext/performance_comparison): | ||
| Especially for bigger grids the functionality can be further optimized by only analysing the outline of a polygon. Here are timings created by calculating the lir for [masks in different resolutions](https://github.com/lukasalexanderweber/lir/tree/main/ext/performance_comparison): | ||
|
|
||
| Timings | Timings (log transformed) | ||
| :-------------------------:|:-------------------------: | ||
| <img width="400" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/performance_comparison/performance_comparison.svg" /> | <img width="400" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/performance_comparison/performance_comparison_log.svg" /> | ||
| 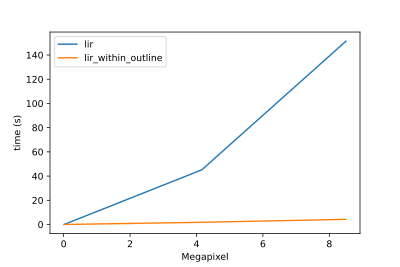 | 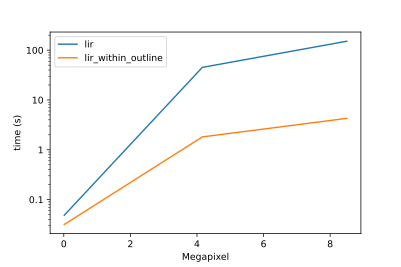 | ||
|
|
||
| The computation costs are saved by analysing only the outline pixels instead of all cells. We utilize the fact that the LIR always touches the outline of the cell grid. Here is how it works: | ||
| The computation costs are saved by analysing only the contour pixels instead of all cells. We utilize the fact that the LIR always touches the outline of the polygon. Here is how it works: | ||
|
|
||
| <img width="200" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/cells2.png"> | ||
|  | ||
|
|
||
| An outline cell can span into one (blue), two (green) or three (red) directions (up, down, left, right): | ||
|
|
||
| <img width="200" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/direction_map.png"> | ||
|  | ||
|
|
||
| By calculating the spans in all possible directions we can obtain a span map: | ||
|
|
||
| Widths | Heights | Areas | ||
| :-------------------------:|:-------------------------:|:-------------------------: | ||
| <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map_widths.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map_heights.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map_areas.png" /> | ||
|  | 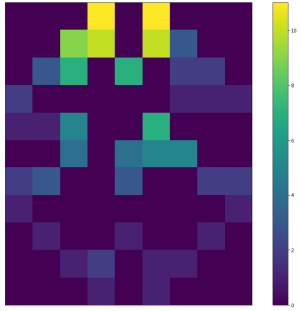 |  | ||
|
|
||
| To analyse what happens here we'll have a closer look at cell (4,2). | ||
| To analyse what happens here we'll have a closer look at cell (4,2). | ||
|
|
||
| <img width="200" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/direction_map_cell_2_2.png"> | ||
| 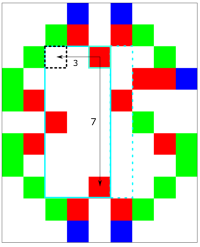 | ||
|
|
||
| It can span into 3 directions: left, down and right. Going to left and down the maximum span is (3 by 7). The final spans are noted in left2right and top2bottom notation. In this case, however, the width is calculated from right2left. We can transform it with the simple formula `x = cell_x - span_width + 1`, in this case `4 - 3 + 1 = 2`. Since the height is already calculated from top2bottom y doesn't change and the span (3 by 7) is allocated to cell (2,2) (black dotted). | ||
|
|
||
| (2,2) is the cell with the biggest area in the span map<sup>1</sup>. However, the information that the rectangle can be expanded to the right (turquoise dotted) is missing. | ||
| (2,2) is (besides (1,6)) the cell with the biggest area in the span map. However, the information that the rectangle can be expanded to the right (turquoise dotted) is missing. | ||
|
|
||
| So for "candidate cells" like (2,2) which do not lie on the outline and come from outline cells going in 3 directions, we create a new span map (using left2right and top2bottom adjacencies): | ||
|
|
||
| <sup>1</sup> TODO cell (1,6) has the same area, there is no feedback to the user if multiple LIRs exist | ||
|
|
||
| <img width="200" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/candidate_map.png"> | ||
|  | ||
|
|
||
| Widths | Heights | Areas | ||
| :-------------------------:|:-------------------------:|:-------------------------: | ||
| <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map2_widths.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map2_heights.png" /> | <img width="300" src="https://github.com/lukasalexanderweber/lir/blob/main/ext/readme_imgs/outline_approach/span_map2_areas.png" /> | ||
| 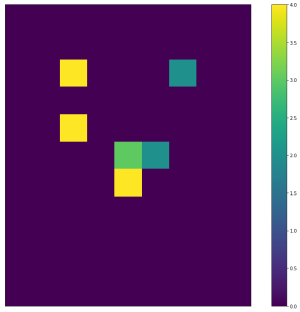 | 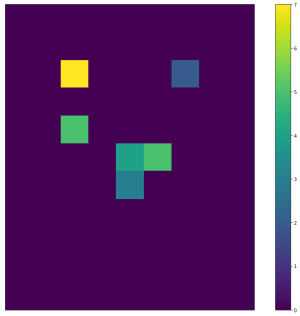 | 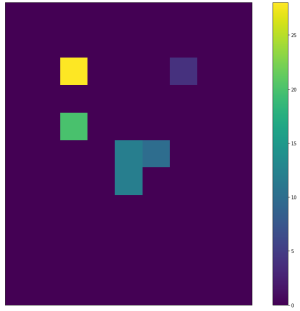 | ||
|
|
||
| The biggest span of the two span maps are compared and the bigger one returned as lir, in this case cell (2,2) with a span (4 by 7) |
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Binary file modified
BIN
+132 KB
(1100%)
ext/readme_imgs/outline_approach/direction_map_cell_2_2.png
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Binary file not shown.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,3 @@ | ||
| from .lir import lir | ||
|
|
||
| __version__ = "0.0.1" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,114 +1,19 @@ | ||
| import numpy as np | ||
| import numba as nb | ||
|
|
||
|
|
||
| def largest_interior_rectangle(cells): | ||
| h_adjacency = horizontal_adjacency(cells) | ||
| v_adjacency = vertical_adjacency(cells) | ||
| s_map = span_map(cells, h_adjacency, v_adjacency) | ||
| return biggest_span_in_span_map(s_map) | ||
|
|
||
|
|
||
| @nb.njit('uint32[:,::1](boolean[:,::1])', parallel=True, cache=True) | ||
| def horizontal_adjacency(cells): | ||
| result = np.zeros((cells.shape[0], cells.shape[1]), dtype=np.uint32) | ||
| for y in nb.prange(cells.shape[0]): | ||
| span = 0 | ||
| for x in range(cells.shape[1]-1, -1, -1): | ||
| if cells[y, x]: | ||
| span += 1 | ||
| else: | ||
| span = 0 | ||
| result[y, x] = span | ||
| return result | ||
|
|
||
|
|
||
| @nb.njit('uint32[:,::1](boolean[:,::1])', parallel=True, cache=True) | ||
| def vertical_adjacency(cells): | ||
| result = np.zeros((cells.shape[0], cells.shape[1]), dtype=np.uint32) | ||
| for x in nb.prange(cells.shape[1]): | ||
| span = 0 | ||
| for y in range(cells.shape[0]-1, -1, -1): | ||
| if cells[y, x]: | ||
| span += 1 | ||
| else: | ||
| span = 0 | ||
| result[y, x] = span | ||
| return result | ||
|
|
||
|
|
||
| @nb.njit('uint32(uint32[:])', cache=True) | ||
| def predict_vector_size(array): | ||
| zero_indices = np.where(array == 0)[0] | ||
| if len(zero_indices) == 0: | ||
| if len(array) == 0: | ||
| return 0 | ||
| return len(array) | ||
| return zero_indices[0] | ||
|
|
||
|
|
||
| @nb.njit('uint32[:](uint32[:,::1], uint32, uint32)', cache=True) | ||
| def h_vector(h_adjacency, x, y): | ||
| vector_size = predict_vector_size(h_adjacency[y:, x]) | ||
| h_vector = np.zeros(vector_size, dtype=np.uint32) | ||
| h = np.Inf | ||
| for p in range(vector_size): | ||
| h = np.minimum(h_adjacency[y+p, x], h) | ||
| h_vector[p] = h | ||
| h_vector = np.unique(h_vector)[::-1] | ||
| return h_vector | ||
|
|
||
|
|
||
| @nb.njit('uint32[:](uint32[:,::1], uint32, uint32)', cache=True) | ||
| def v_vector(v_adjacency, x, y): | ||
| vector_size = predict_vector_size(v_adjacency[y, x:]) | ||
| v_vector = np.zeros(vector_size, dtype=np.uint32) | ||
| v = np.Inf | ||
| for q in range(vector_size): | ||
| v = np.minimum(v_adjacency[y, x+q], v) | ||
| v_vector[q] = v | ||
| v_vector = np.unique(v_vector)[::-1] | ||
| return v_vector | ||
|
|
||
|
|
||
| @nb.njit('uint32[:,:](uint32[:], uint32[:])', cache=True) | ||
| def spans(h_vector, v_vector): | ||
| spans = np.stack((h_vector, v_vector[::-1]), axis=1) | ||
| return spans | ||
|
|
||
|
|
||
| @nb.njit('uint32[:](uint32[:,:])', cache=True) | ||
| def biggest_span(spans): | ||
| if len(spans) == 0: | ||
| return np.array([0, 0], dtype=np.uint32) | ||
| areas = spans[:, 0] * spans[:, 1] | ||
| biggest_span_index = np.where(areas == np.amax(areas))[0][0] | ||
| return spans[biggest_span_index] | ||
|
|
||
|
|
||
| @nb.njit('uint32[:, :, :](boolean[:,::1], uint32[:,::1], uint32[:,::1])', | ||
| parallel=True, cache=True) | ||
| def span_map(cells, h_adjacency, v_adjacency): | ||
|
|
||
| y_values, x_values = cells.nonzero() | ||
| span_map = np.zeros(cells.shape + (2,), dtype=np.uint32) | ||
|
|
||
| for idx in nb.prange(len(x_values)): | ||
| x, y = x_values[idx], y_values[idx] | ||
| h_vec = h_vector(h_adjacency, x, y) | ||
| v_vec = v_vector(v_adjacency, x, y) | ||
| s = spans(h_vec, v_vec) | ||
| s = biggest_span(s) | ||
| span_map[y, x, :] = s | ||
|
|
||
| return span_map | ||
|
|
||
|
|
||
| @nb.njit('uint32[:](uint32[:, :, :])', cache=True) | ||
| def biggest_span_in_span_map(span_map): | ||
| areas = span_map[:, :, 0] * span_map[:, :, 1] | ||
| largest_rectangle_indices = np.where(areas == np.amax(areas)) | ||
| x = largest_rectangle_indices[1][0] | ||
| y = largest_rectangle_indices[0][0] | ||
| span = span_map[y, x] | ||
| return np.array([x, y, span[0], span[1]], dtype=np.uint32) | ||
| from .lir_basis import largest_interior_rectangle as lir_basis | ||
| from .lir_within_contour import largest_interior_rectangle \ | ||
| as lir_within_contour | ||
|
|
||
|
|
||
| def lir(grid, contour=None): | ||
| """ | ||
| Returns the Largest Interior Rectangle of a binary grid. | ||
| :param grid: 2D ndarray containing data with `bool` type. | ||
| :param contour: (optional) 2D ndarray with shape (n, 2) containing | ||
| xy values of a specific contour where the rectangle could start | ||
| (in all directions). | ||
| :return: 1D ndarray with lir specification: x, y, width, height | ||
| :rtype: ndarray | ||
| """ | ||
| if contour is None: | ||
| return lir_basis(grid) | ||
| else: | ||
| return lir_within_contour(grid, contour) |
Oops, something went wrong.