Department of Engineering Information, University of Padua
Pietrobon Andrea
•
Biffis Nicola
Explore the docs »
View Demo on Google Colab
•
Report Bug
This project was tested on an NVIDIA GeForce GTX 1050 Ti with 4GB of memory, as well as with the Google Colab free plan. Consequently, it had limited usage of a GPU. This allowed us to achieve the following results, which, unfortunately, in our opinion, are not entirely satisfactory. However, we are confident that with the use of a more powerful GPU, better results can be obtained. For this reason, we encourage anyone who wants to try using GPUs with significantly more memory available.
This research will show an innovative method useful in the segmentation of polyps during the screening phases of colonoscopies with the aim of concretely helping doctors in this task. To do this we have adopted a new approach which consists in merging the hybrid semantic network (HSNet) architecture model with the Reagion-wise(RW) as a loss function for the backpropagation process. In this way the bottleneck problems that arise in the systems currently used in this area are solved, since thanks to the HSNet it is possible to exploit the advantages of both the Transformers and the convolutional neural networks and thanks to the RW loss function its capacity is exploited to work efficiently with biomedical images. Since both the architecture and the loss function chosen by us have shown that they can achieve performances comparable to those of the state of the art working individually, in this research a dataset divided into 5 subsets will be used to demonstrate their effectiveness by using them together .
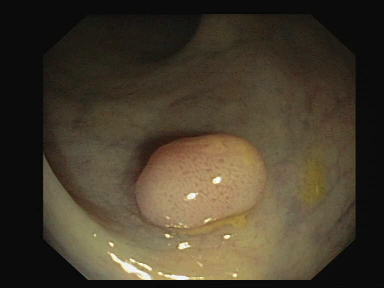
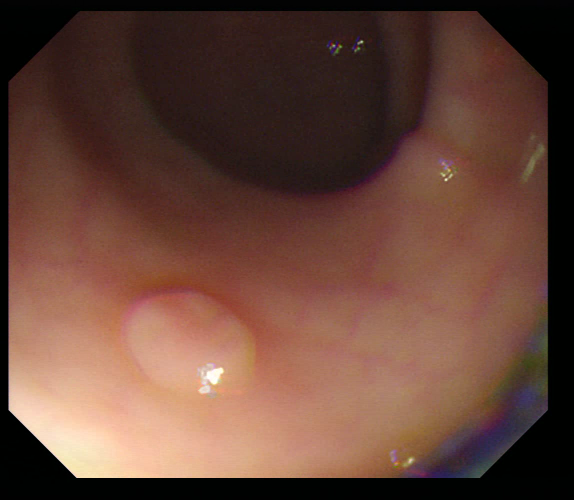
| Original Image | True Mask | Output |
|---|---|---|
 |
 |
 |
 |
 |
 |
This project was built using a variety of technologies like: Python 3.9, renowned for its versatility and readability; PyTorch, revered for its prowess in deep learning applications; OpenCV, an indispensable tool for computer vision tasks; Numpy, revered for its array computing capabilities; along with a plethora of other third party libraries like nnUNet, that collectively contribute to the project's robustness and functionality.
In the field of biomedicine, quantitative analysis requires a crucial step: image segmentation. Manual segmentation is a time-consuming and subjective process, as demonstrated by the considerable discrepancy between segmentations performed by different annotators. Consequently, there is a strong interest in developing reliable tools for automatic segmentation of medical images.
The use of neural networks to automate polyp segmentation can provide physicians with an effective tool for identifying such formations or areas of interest during clinical practice. However, there are two important challenges that limit the effectiveness of this segmentation:
- Polyps can vary significantly in size, orientation, and illumination, making accurate segmentation difficult to achieve.
- Current approaches often overlook significant details such as textures.
To obtain precise segmentations in medical imagesegmentation, it is crucial to consider class imbalance and the importance of individual pixels. By pixel importance, we refer to the phenomenon where the severity of classification errors depends on the position of such errors.
Current approaches for polyp segmentation primarily rely on convolutional neural networks (CNN)
or Transformers, so to overcome the mentioned challenges, this research proposes the use of a hybrid semantic network (HSNet) that combines the advantages of Transformer networks and convolutional neural networks (CNN), along with regional loss (RW). Thanks to this loss function we can simultaneously takes into account class imbalance and pixel importance, without requiring additional hyperparameters or functions, in order to improve polyp segmentation.
In this study, we examine how the implementation of regional loss (RW) affects polyp segmentation by applying it to a hybrid semantic network (HSNet).
Read the paper for a more detailed explanation: Documentation »
This are the dependencies you need to test the project. It's strongly suggested to use python 3.9 (or higher).
pip3 install thop
pip3 install libtiff
pip3 install timm
pip3 install opencv-python
pip3 install scipy
pip3 install numpy
pip3 install nibabel
pip3 install nnunet
pip3 install matplotlib
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu117You can download the dataset from this Google Drive Link
Those are the steps to follow to test the code. Note that you need a NVIDIA GPU to run the code. If when you start the code you get an error like this:
torch.cuda.OutOfMemoryError: CUDA out of memory. Tried to allocate 20.00 MiB (GPU 0;
4.00 GiB total capacity; 3.36 GiB already allocated; 0 bytes free; 3.48 GiB reserved
in total by PyTorch) If reserved memory is >> allocated memory try setting
max_split_size_mb to avoid fragmentation.See documentation for Memory Management
and PYTORCH_CUDA_ALLOC_CONF
It means that you don't have enough memory on your GPU. For this reason a GPU with at least 8GB of memory is recommended. Alternatively you can test it on Google Colab from HERE.
-
Clone the repo
git clone https://github.com/Piero24/lossFuncition-4SN.git
-
Download the dataset from Google Drive
-
Change the paths of the dataset in
folder_path.py -
Check the hyperparameters.
The hyperparameters used in our tests are the followings:
a) learning rate: 0.0001
b) training size: 160
c) batch size: 24
d) number of epochs: 50⚠️ ATTENTION: Be careful when changing the training size and batch size. For an unclear problem (maybe internal to pytorch) we can use multiples of 8 up to 352 for the training size and the batch size must be a dividend of the training size. -
Start the net with
python main/Train.pyNOTE: At the end of each epoch the test will be carried out.
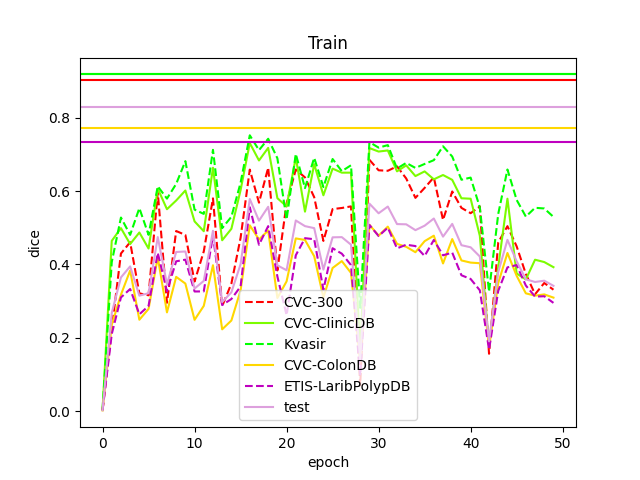
The model for each test will be saved.Every 10 epochs the dice plot for the test phase is saved (Each plot starts from the very first epoch up to the current number, for example 0-10, 0-20 ... 0-100). At the same time a table is saved with the actual dice value for the last 10 test sets (e.g. 0-10, 10-20 ... 90-100).
Once this phase has been completed, each saved model will be tested on a portion of the test set and will save the relative masks in order to be able to make a visual comparison.
We assume no responsibility for an improper use of this code and everything related to it. We do not assume any responsibility for damage caused to people and / or objects in the use of the code.
By using this code even in a small part, the developers are declined from any responsibility.It is possible to have more information by viewing the following links: Code of conduct • License
MIT License
Copyright (c) 2023 Andrea Pietrobon
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
License Documentation »
The individual licenses are indicated in the following section.
Software list:
| Software | License owner | License type |
|---|---|---|
| HSNet | baiboat | * |
| HarDNet-MSEG | james128333 | Apache-2.0 |
| Polyp-PVT | DengPingFan | * |
| Region-wise Loss for Biomedical Image Segmentation | Juan Miguel Valverde, Jussi Tohka | |
| pytorch-image-models | huggingface | Apache-2.0 |
| thop | Lyken17 | MIT |
| nnUNet | MIC-DKFZ | Apache-2.0 |
Copyrright (C) by Pietrobon Andrea
Released date: July-2023