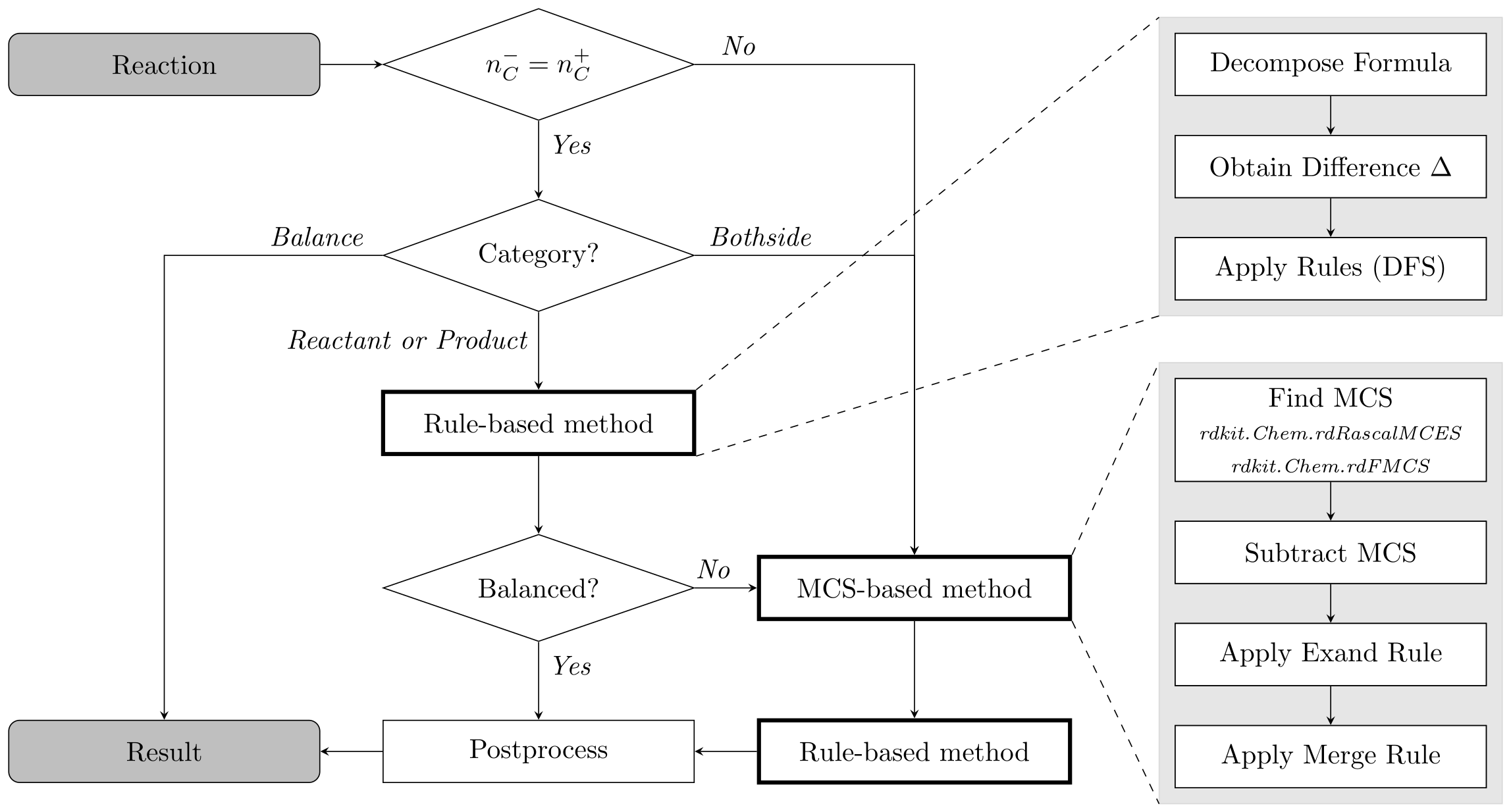
SynRBL is a toolkit tailored for computational chemistry, aimed at correcting imbalances in chemical reactions. It employs a dual strategy: a rule-based method for adjusting non-carbon elements and an mcs-based (maximum common substructure) technique for carbon element adjustments.
The easiest way to use SynRBL is by installing the PyPI package synrbl.
Follow these steps to setup a working environment. Please ensure you have Python 3.11 or later installed on your system.
The requirements are automatically installed with the pip package.
- Python 3.11
- rdkit >= 2023.9.4
- joblib >= 1.3.2
- seaborn >= 0.13.2
- xgboost >= 2.0.3
- scikit_learn == 1.4.0
- imbalanced_learn >= 0.12.0
- reportlab >= 4.1.0
- fgutils >= 0.1.3
-
Python Installation: Ensure that Python 3.11 or later is installed on your system. You can download it from python.org.
-
Creating a Virtual Environment (Optional but Recommended): It's recommended to use a virtual environment to avoid conflicts with other projects or system-wide packages. Use the following commands to create and activate a virtual environment:
python -m venv synrbl-env
source synrbl-env/bin/activate # On Windows use `synrbl-env\Scripts\activate`Or Conda
conda create --name synrbl-env python=3.11
conda activate synrbl-env- Install with pip:
pip install synrbl- Verify Installation: After installation, you can verify that SynRBL is correctly installed by running a simple test.
python -c "from synrbl import Balancer; bal = Balancer(n_jobs=1); print(bal.rebalance('CC(=O)OCC>>CC(=O)O'))"from synrbl import Balancer
smiles = (
"COC(=O)[C@H](CCCCNC(=O)OCc1ccccc1)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O>>"
+ "COC(=O)[C@H](CCCCN)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O"
)
synrbl = Balancer()
results = synrbl.rebalance(smiles, output_dict=True)
>> [{
"reaction": "COC(=O)[C@H](CCCCNC(=O)OCc1ccccc1)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O.O>>"
+ "COC(=O)[C@H](CCCCN)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O.O=C(O)OCc1ccccc1",
"solved": True,
"input_reaction": "COC(=O)[C@H](CCCCNC(=O)OCc1ccccc1)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O>>"
+ "COC(=O)[C@H](CCCCN)NC(=O)Nc1cc(OC)cc(C(C)(C)C)c1O",
"issue": "",
"rules": ["append O when next to O or N", "default single bond"],
"solved_by": "mcs-based",
"confidence": 0.999,
}]echo "id,reaction\n0,CC(=O)OCC>>CC(=O)O" > unbalanced.csv
python -m synrbl run -o balanced.csv unbalanced.csvPrepare your dataset as a csv file datafile with a column reaction of unbalanced reaction SMILES and a column expected_reaction containing the expected balanced reactions.
Rebalance the reactions and forward the expected reactions column to the output.
python -m synrbl run -o balanced.csv --col <reaction> --out-columns <expected_reaction> <datafile>After rebalancing you can use the benchmark command to compute the success and accuracy rates of your dataset. Keep in mind that an exact comparison between rebalanced and expected reaction is a highly conservative evaluation. An unbalance reaction might have multiple equaly viable balanced solutions. Besides the exact comparison (default) the benchmark command supports a few similarity measures like ECFP and pathway fingerprints for the comparison between rebalanced reaction and the expected balanced reaction.
python -m synrbl benchmark --col <reaction> --target-col <expected_reaction> balanced.csvTo test SynRBL on the provided validation set use the following commands. Run these commands from the root of the cloned repository.
Rebalance the dataset
python -m synrbl run -o validation_set_balanced.csv --out-columns expected_reaction ./Data/Validation_set/validation_set.csvand compute the benchmark results
python -m synrbl benchmark validation_set_balanced.csvThis project is licensed under MIT License - see the License file for details.
Reaction rebalancing: a novel approach to curating reaction databases
@Article{Phan2024,
author={Phan, Tieu-Long and Weinbauer, Klaus and G{\"a}rtner, Thomas and Merkle,
Daniel and Andersen, Jakob L. and Fagerberg, Rolf and Stadler, Peter F.},
title={Reaction rebalancing: a novel approach to curating reaction databases},
journal={Journal of Cheminformatics},
year={2024},
month={Jul},
day={19},
volume={16},
number={1},
pages={82},
issn={1758-2946},
doi={10.1186/s13321-024-00875-4},
url={https://doi.org/10.1186/s13321-024-00875-4}
}
This project has received funding from the European Unions Horizon Europe Doctoral Network programme under the Marie-Skłodowska-Curie grant agreement No 101072930 (TACsy -- Training Alliance for Computational)