This package computes exact solutions to the
The algorithm works as follows:
- We run
$k$ -means++ on the data to get an initial upper bound for the cost of an optimal solution. - We try to find the largest cluster size
$c_1$ that a cluster in the optimal solution may have. We use the fact that the cost of a cluster in the optimal solution is less than the initial upper bound. We do this by solving an ILP (ILP#1). - We iterate through all possible cluster sizes
$c_1 \geq c_2 \ldots \geq c_k$ and compute for each of them a solution using an ILP (ILP#2). However, we don't actually iterate through all the sizes, but use a branch-and-bound algorithm to skip some of them if the cost exceeds the initial upper bound. You can see an example of this in the tree in the "Plot the Branch and Bound Tree" section.
Both ILPs get cluster sizes
- the number of clusters is
$i$ - the clusters have sizes
$c_1,\ldots, c_i$ - if
$n$ is the number of data points$n-c_1-\ldots -c_i$ points are not part of any cluster
ILP#1 gets one single cluster size
ILP#2 gets cluster sizes
To search for the cluster sizes of an optimal solution we use a branch and bound approach. In a branch node of level branching_levels and fill_cluster_sizes define the behavior on these branching nodes. If branching_levels is greater equal to the level fill_cluster_sizes is set to true we compute the smallest possible remaining cluster sizes fill_cluster_sizes is set to false we run ILP#2 only with the fixed cluster sizes fill_cluster_sizes to true may lead to less branching but can increase the solving time of ILP#2.
To customize the runs, you can create a config file. The default config file is exact_kmeans/config/default.yaml. You can also pass a different config file as an argument.
-
num_processes(integer or float) sets the number of processes used. The algorithm was parallelized using themultiprocessingpackage, so you can set the number of processes that you want to use. If you use an integer, at most that number of processes will be taken, otherwise if you use a float, it will be a fraction of the available CPUs. If the parameter is not passed, the algorithm will use all available CPUs. -
bound_model_paramsare the arguments that are passed to the ILP#1 model. Please have a look at the Gurobi documentation for more information. -
model_paramsare the arguments that are passed to the ILP#2 model. Please have a look at the Gurobi documentation for more information. -
branching_priorities(true/false) enables the use of branching priorities. If true, the priority of the$x$ variable will be higher than the other variables. According to our tests, this speeds up the solving. -
replace_min(true/false) replaces a minimum constraint with a linear version. According to our tests, this speeds up the solving. -
branching_levels(integer) Sets the maximum level of a branching node where we run ILP#2 to bound the cost. Since this can increase the number of ILPs that are computed, it may only make sense for small levels. -
fill_cluster_sizes(true/false) If set to false we run ILP#2 only with cluster sizes fixed at the branching node. If set to true we fill the cluster sizes up to$k$ . Setting the variable to true can result in a larger computation time for ILP#2 but may result in less branching, which can save time. For small$k$ we recommend setting it to false and for large$k$ we recommend setting it to true.
For this package to work, you need to have a working installation of Gurobi. You can get a free academic license here. Here you can find a guide on how to install it.
pip install exact-kmeansfrom exact_kmeans import ExactKMeans
from exact_kmeans.util import JsonEncoder
from ucimlrepo import fetch_ucirepo
# For example, you can import data from the UCI repository
# but you can also use your own dataset
iris = fetch_ucirepo(id=53)
X = iris.data.features
ilp = ExactKMeans(n_clusters=3)
res = ilp.fit(X)
with open("output.json", "w") as f:
json.dump(res, f, indent=4, cls=JsonEncoder)
# You can also print the branch and bound tree to visualize the decisions made by the algorithm
# Below you find an example of the tree and meaning of the colors
from exact_kmeans.plot_tree import plot
plot(
nodes=res.processed_cluster_sizes,
filename="test",
plot_folder="plots",
optimal_objective=res.model.ObjVal,
)Install poetry
curl -sSL https://install.python-poetry.org | python3 -Install the package
poetry installRun the program
poetry run python -m exact_kmeans --data-path iris.csv --verbose --results-path test/iris.json --k 3 --config-file exact_kmeans/config/default.yamlYour data-path should be a file with a header containing only the comma-separated data points. Example:
0,1,2,3
5.1,3.5,1.4,0.2
4.9,3.0,1.4,0.2
4.7,3.2,1.3,0.2
4.6,3.1,1.5,0.2
5.0,3.6,1.4,0.2
...
Assuming that you have run the program and stored the results in a JSON file, you can plot the tree produced by the algorithm.
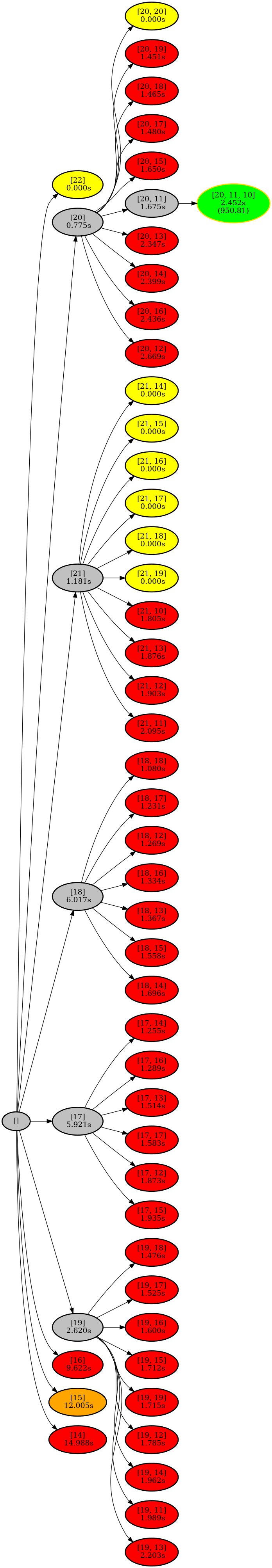
poetry run python -m exact_kmeans.plot_tree --output-json output.json --plot-folder plotsThis will create a tree that looks like this:
The nodes can have the following colors:
- Red: The cluster sizes were infeasible.
- Green: The cluster sizes were feasible.
- Gray: The node was a branching node.
- Orange/Yellow: The node was skipped because it had a too high initial cost.
- The optimal node has a gold border.
The nodes have the following information:
- The cluster sizes that were tested.
- The amount of seconds it took.
- The cost of the solution. This is only shown if the node has
$k$ clusters and is feasible.
Install poetry
curl -sSL https://install.python-poetry.org | python3 -Install the package
poetry installRun the tests
poetry run python -m unittest discover tests -vThis package requires the Gurobi Optimizer, which is a commercial optimization solver. Users must obtain their own licenses for Gurobi. You are responsible for complying with all Gurobi licensing terms. For more information, visit the Gurobi Licensing Page.