PPIRef is a Python package for working with 3D structures of protein-protein interactions (PPIs). It is based on the PPIRef dataset, comprising all PPIs from the Protein Data Bank (PDB). The package aims to provide standard data and tools for machine learning and data science applications involving protein-protein interaction structures. PPIRef includes the following functionalities:
- ⭐ Extracting protein-protein interfaces from .pdb files.
- ⭐ Visualizing and analyzing the properties of PPIs.
- ⭐ Comparing, deduplicating and clustering PPI interfaces.
- ⭐ Retrieving similar PPIs from PDB by similar interface structure or sequence.
- ⭐ Downloading, splitting and subsampling prepared PPIs for machine learning applications.
Please see the documentation for usage examples and API reference. See also our paper for additional details.
Install the PPIRef package.
conda create -n ppiref python=3.10
conda activate ppiref
git clone https://github.com/anton-bushuiev/PPIRef.git
cd PPIRef; pip install -e .Download the dataset using the package (in Python).
from ppiref.utils.misc import download_from_zenodo
from ppiref.split import read_fold
from ppiref.utils.ppi import PPI
download_from_zenodo('ppi_6A.zip') # or for example 'pdb_redo_ppi_10A.zip' for all 10-Angstrom PPIs from PDB-REDO
> Downloading: 100%|██████████| 6.94G/6.94G [10:19<00:00, 11.2MiB/s]
> Extracting: 100%|██████████| 831382/831382 [02:36<00:00, 5313.49files/s]Read the data fold/subset you need (whole PPIRef50K in the example).
ppi_paths = read_fold('ppiref_6A_filtered_clustered_04', 'whole')
print('Dataset size:', len(ppi_paths))
> Dataset size: 51755Now you are ready to work with the PPIRef dataset! Example of a sample:
ppi = PPI(ppi_paths[0])
print('Path:', ppi.path)
print('Statistics:', ppi.stats)
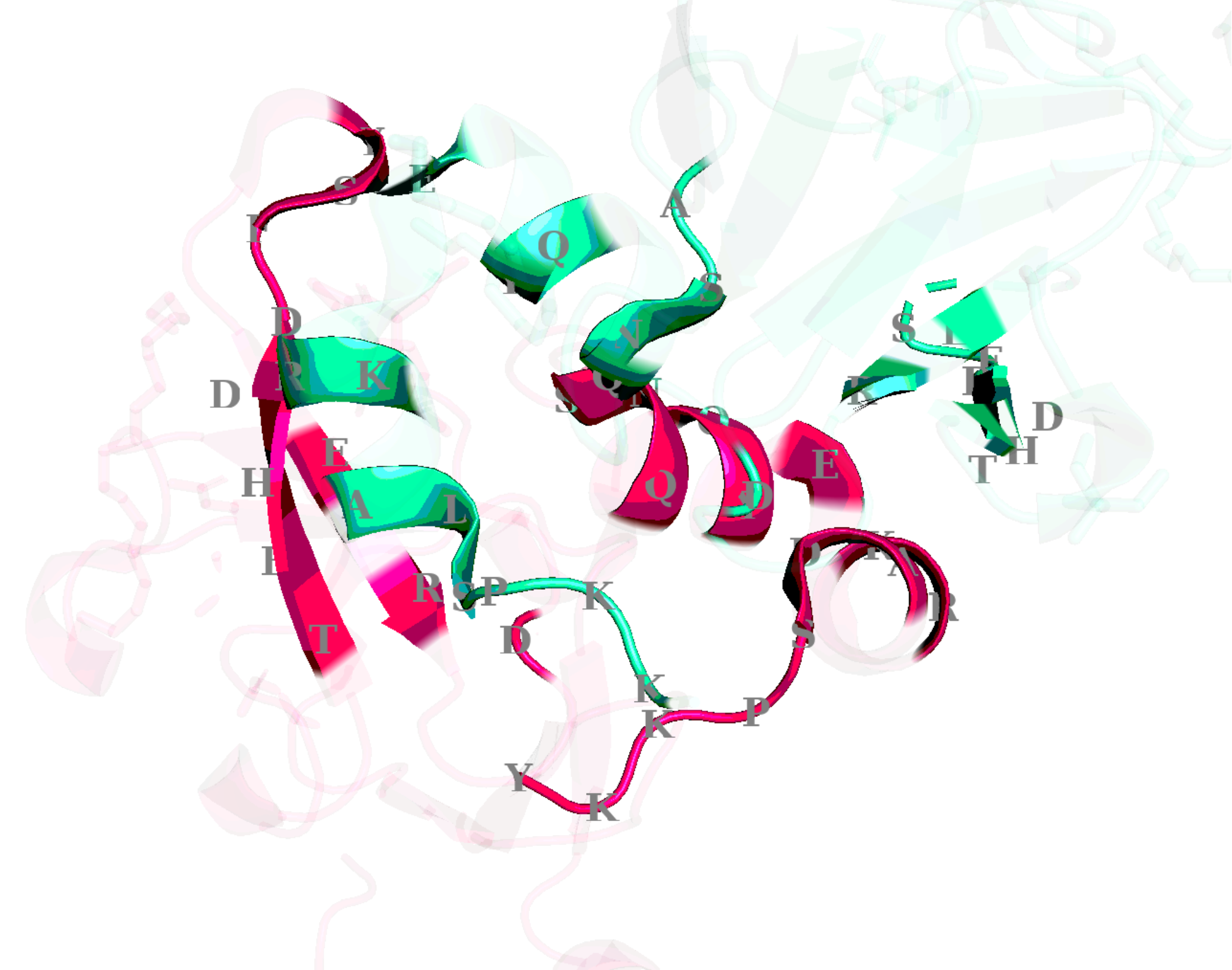
ppi.visualize()
> Path: /Users/anton/dev/PPIRef/ppiref/data/ppiref/ppi_6A/hc/3hch_A_B.pdb
> Statistics:
> {'KIND': 'heavy',
> 'EXTRACTION RADIUS': 6.0,
> 'EXPANSION RADIUS': 0.0,
> 'RESOLUTION': 2.1,
> 'STRUCTURE METHOD': 'x-ray diffraction',
> 'DEPOSITION DATE': '2009-05-06',
> 'RELEASE DATE': '2009-10-13',
> 'BSA': 682.5337386399999}Further, the PPIRef package provides utilities for comparing, deduplicating, and clustering PPI interfaces, as well as for retrieving similar PPIs from PDB by similar interface structure or sequence. Please see the documentation for more details.
The repository is under development. Please do not hesitate to contact us or create an issue/PR if you have any questions or suggestions ✌️.
Technical
- PPIRef (6A interfaces) on Zenodo
- PPIRef (10A interfaces) on Zenodo (expected in June 2024)
- PPIRef version based on the PDB-REDO database for higher-quality side chains in the structures (expected in June 2024)
- Docstrings
Enhancements
- Cluster all PPIs to sample from clusters rather than removing near duplicates completely (similar to UniRef seeds)
- Add RASA values to classify residues according to Levy 2010
- Classify PPIs according to Ofran2003
If you find this repository useful, please cite our paper:
@article{bushuiev2024learning,
title={Learning to design protein-protein interactions with enhanced generalization},
author={Anton Bushuiev and Roman Bushuiev and Petr Kouba and Anatolii Filkin and Marketa Gabrielova and Michal Gabriel and Jiri Sedlar and Tomas Pluskal and Jiri Damborsky and Stanislav Mazurenko and Josef Sivic},
booktitle={ICLR 2024 (The Twelfth International Conference on Learning Representations)},
url={https://doi.org/10.48550/arXiv.2310.18515},
year={2024}
}If relevant, please also cite the corresponding paper on data leakage in protein interaction benchmarks:
@article{bushuiev2024revealing,
title={Revealing data leakage in protein interaction benchmarks},
author={Anton Bushuiev and Roman Bushuiev and Jiri Sedlar and Tomas Pluskal and Jiri Damborsky and Stanislav Mazurenko and Josef Sivic},
booktitle={ICLR 2024 Workshop on Generative and Experimental Perspectives for Biomolecular Design},
url={https://doi.org/10.48550/arXiv.2404.10457},
year={2024}
}If you find any of the external software useful, please cite the corresponding papers (see PPIRef/external/README.md).