This repository contains code, resources, and tutorials described in [1].
| [1] | Hoyt, C. T., Domingo-Fernández, D., & Hofmann-Apitius, M. (2018). BEL Commons: an environment for exploration and analysis of networks encoded in Biological Expression Language. Database, 2018(3), 1–11. |
- data: several folders containing instructions how to automatically download and process differential gene expression experiments
- screenshots: the screenshots for the tutorial (see below)
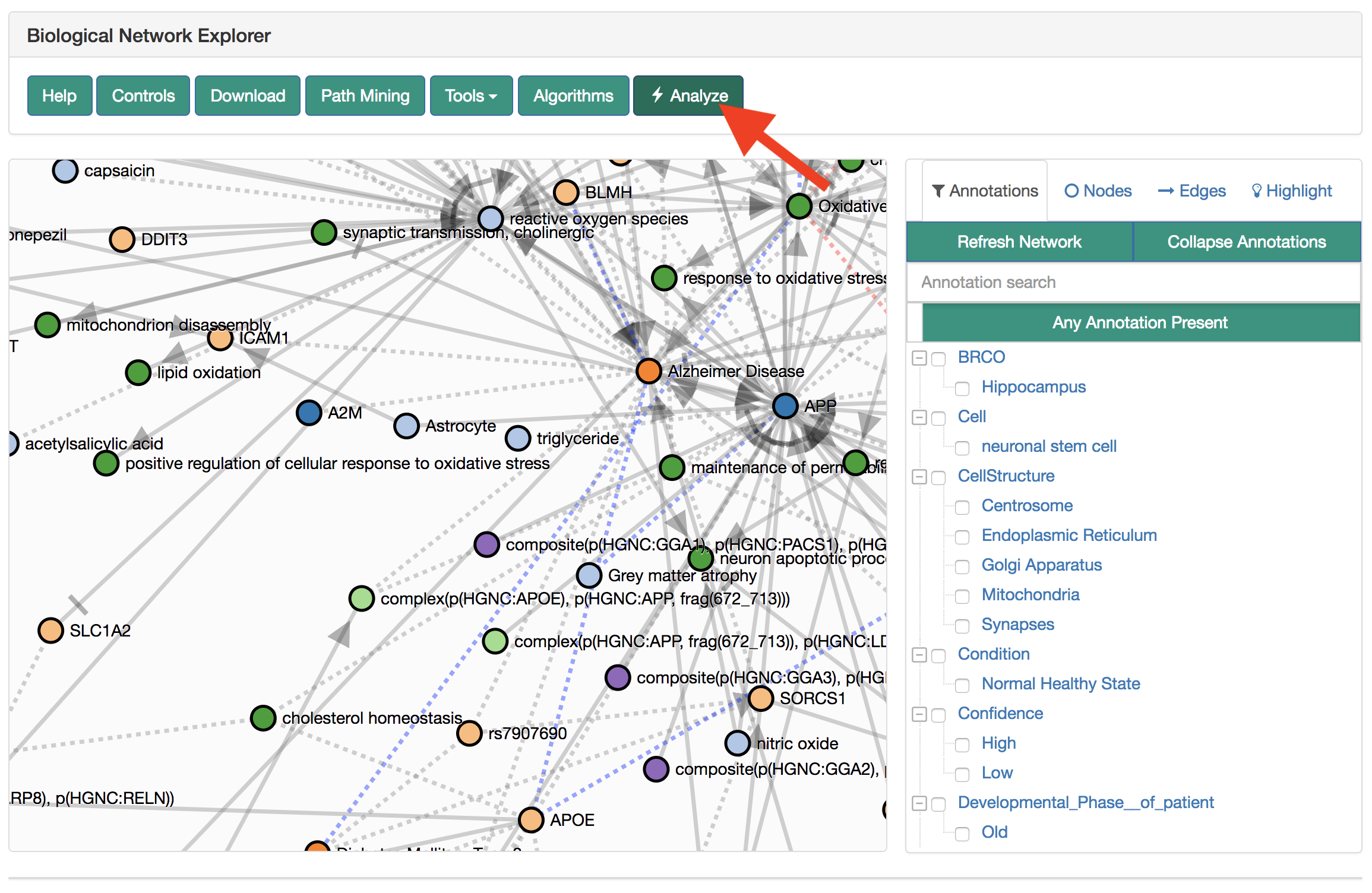
This tutorial shows how to reproduce the analysis shown in Figure 5 of the manuscript.
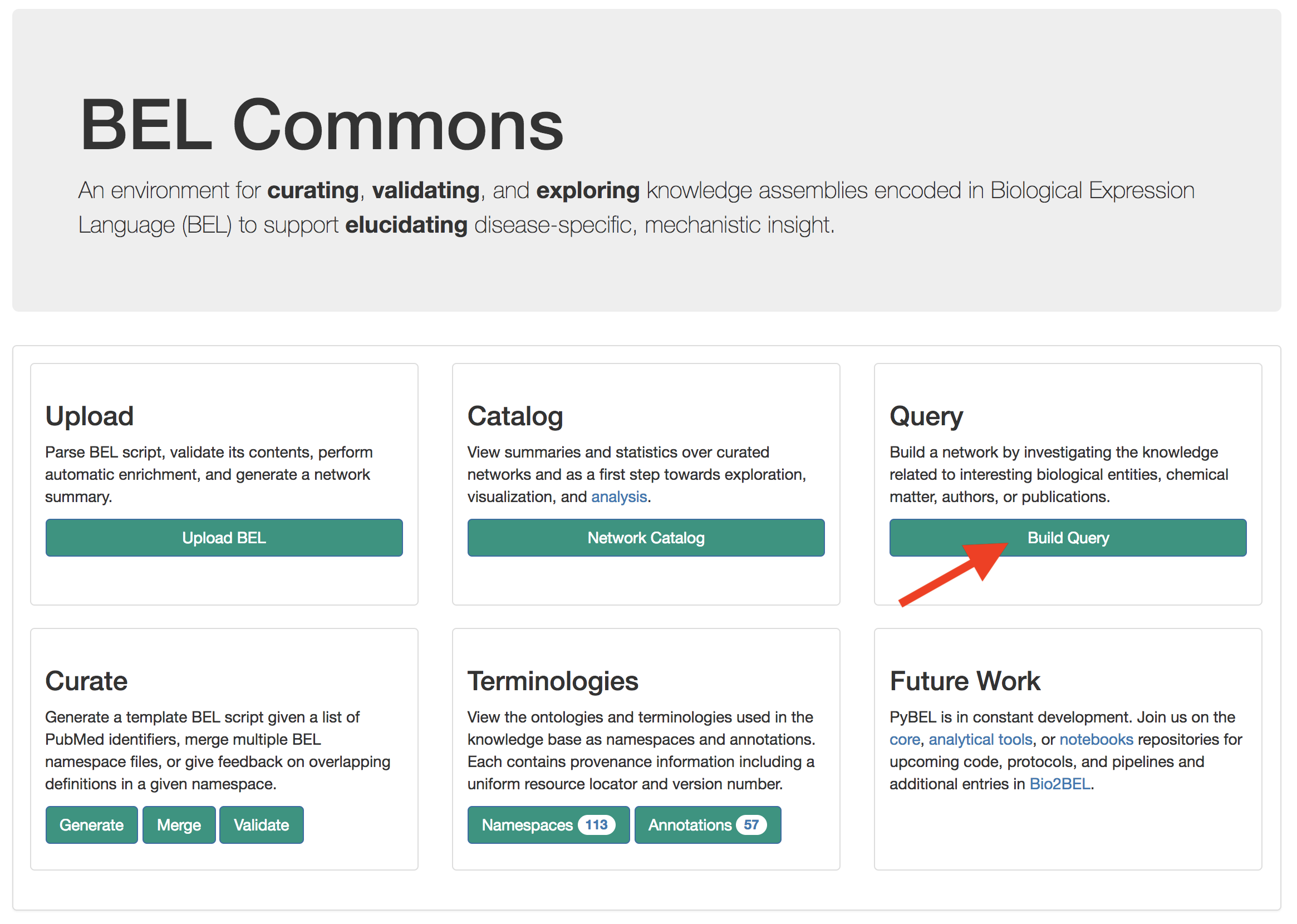
From the main page on BEL Commons, find the query builder.
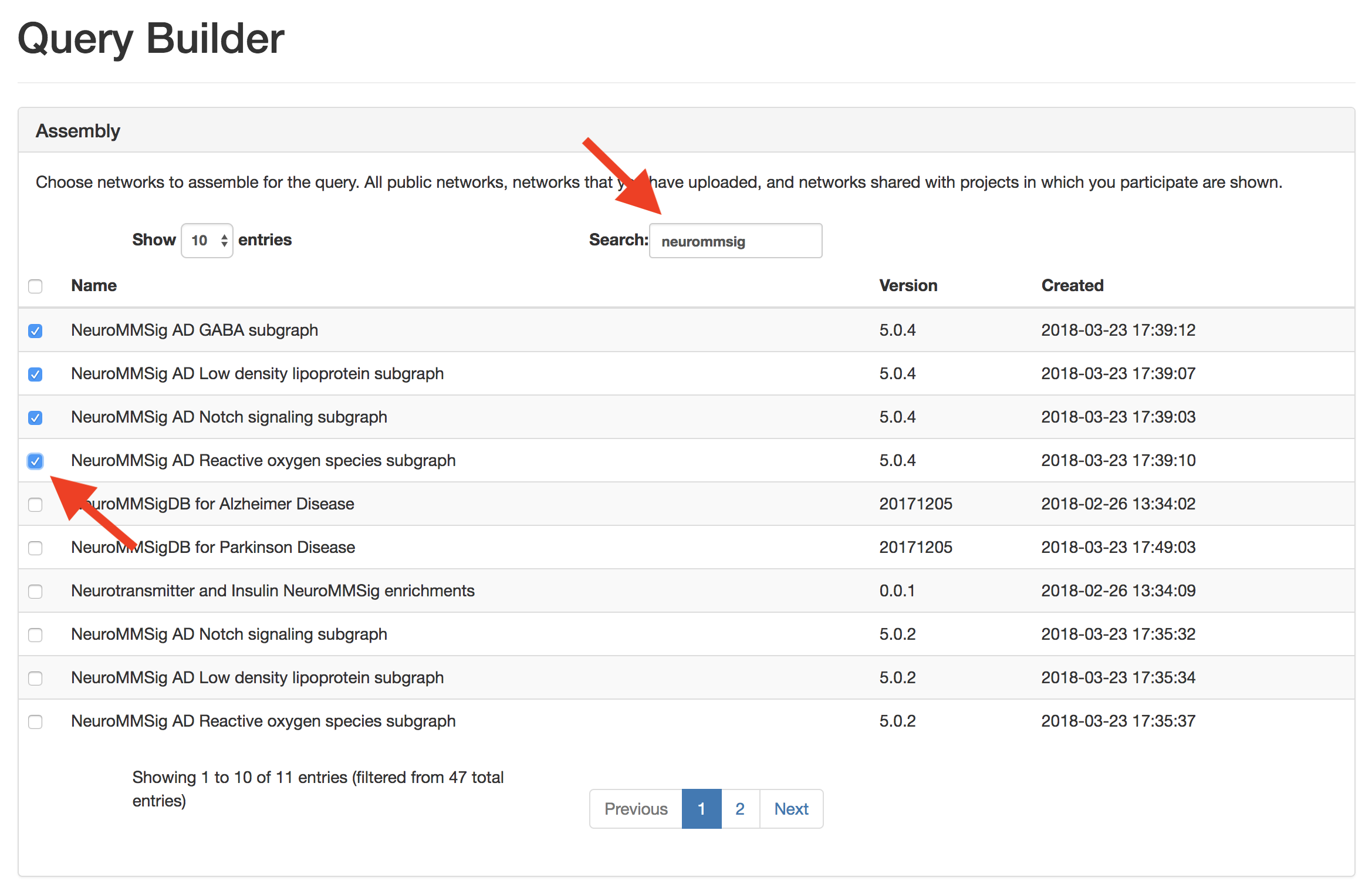
Use the search box to find the example NeuroMMSig AD subgraphs. Click their radio boxes to include in the query
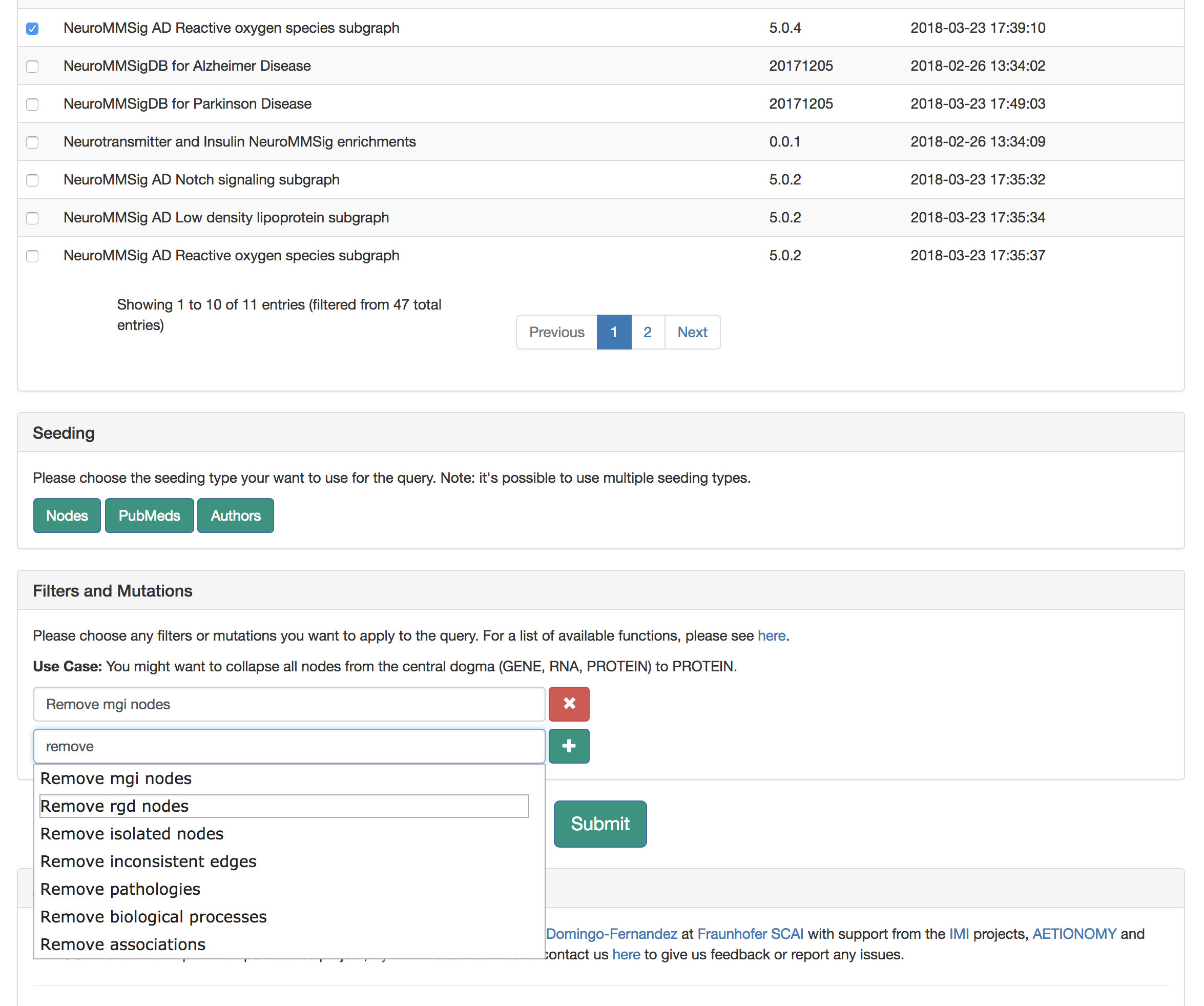
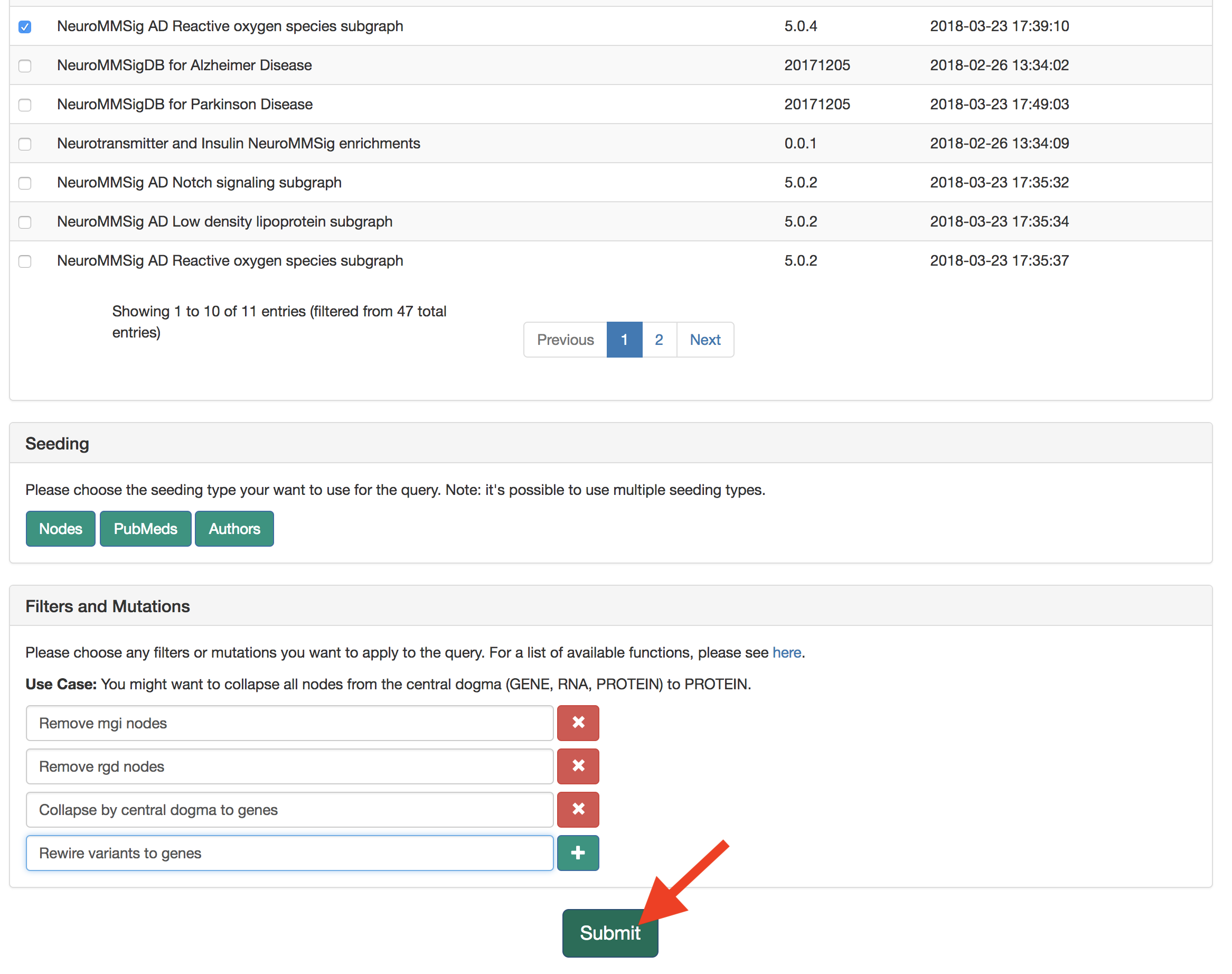
This query does not need any seeding, so scroll down to "Filters and Mutations." As a pre-processing step, use the "Filters and Mutations" box to add functions to remove MGI nodes, RGD nodes, collapse on central dogma, and rewire variants.
Submit the query
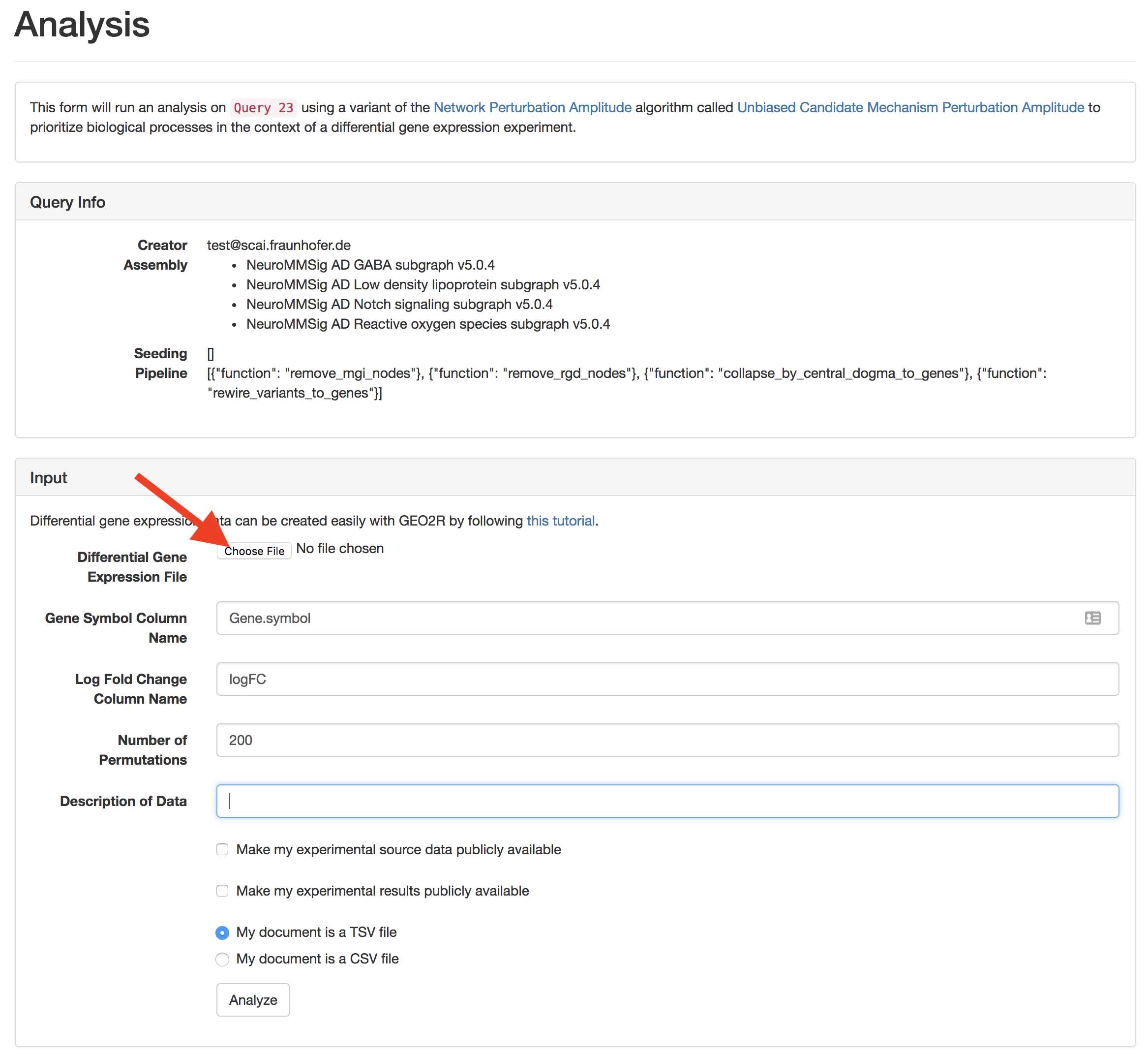
Click the "analyze" button to go to the data upload page
Upload a differential gene expression file. Several examples are included in this repository. In the manuscript, we present an analysis on the three results from GSE28146. Select the number of permutations (more are better since it's a randomized algorithm, but it's much slower. A good number is 500.). Finally, add a description so it's easier to identify the data later and press the submit button.
If the data is malformed, you'll get a warning.
After submitting, you'll be redirected to the same page where another experiment can be run on the same query. This is useful for making differential analysis on the same network with different data sets, as presented in the manuscript. Submit several other experiments, and wait for them to complete.
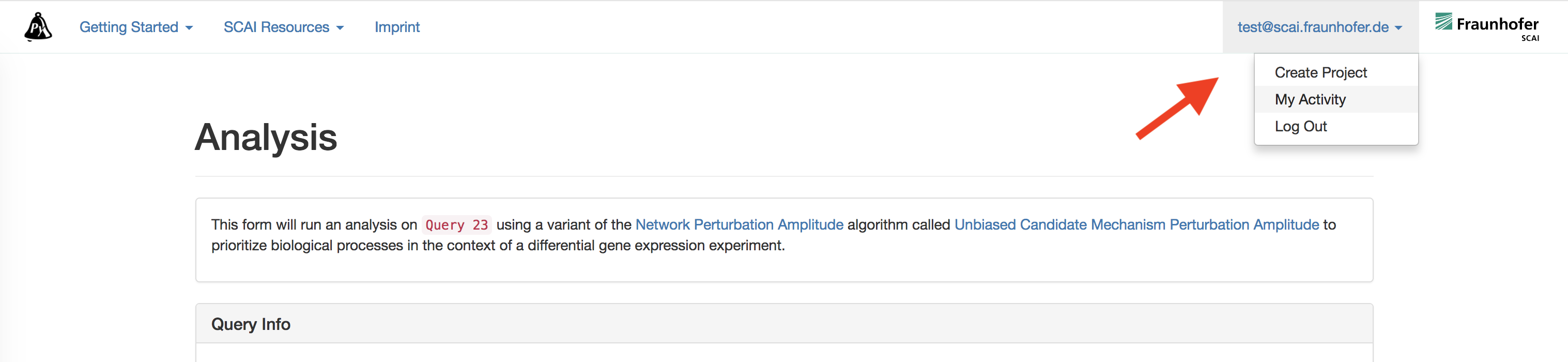
Navigate to "My Activity" from the top of the browser
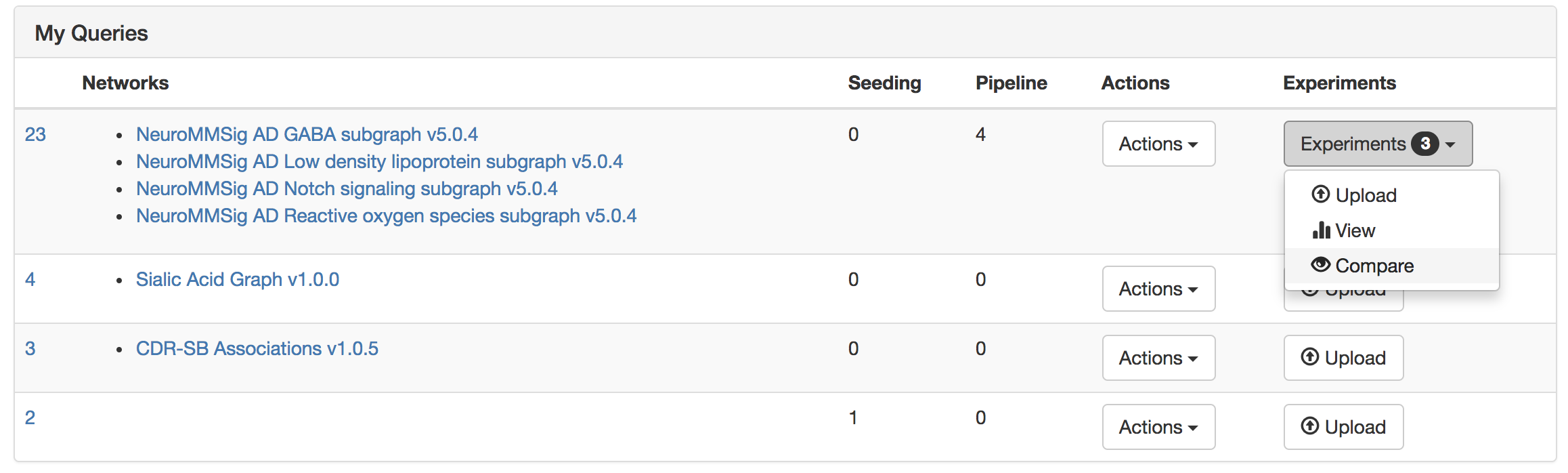
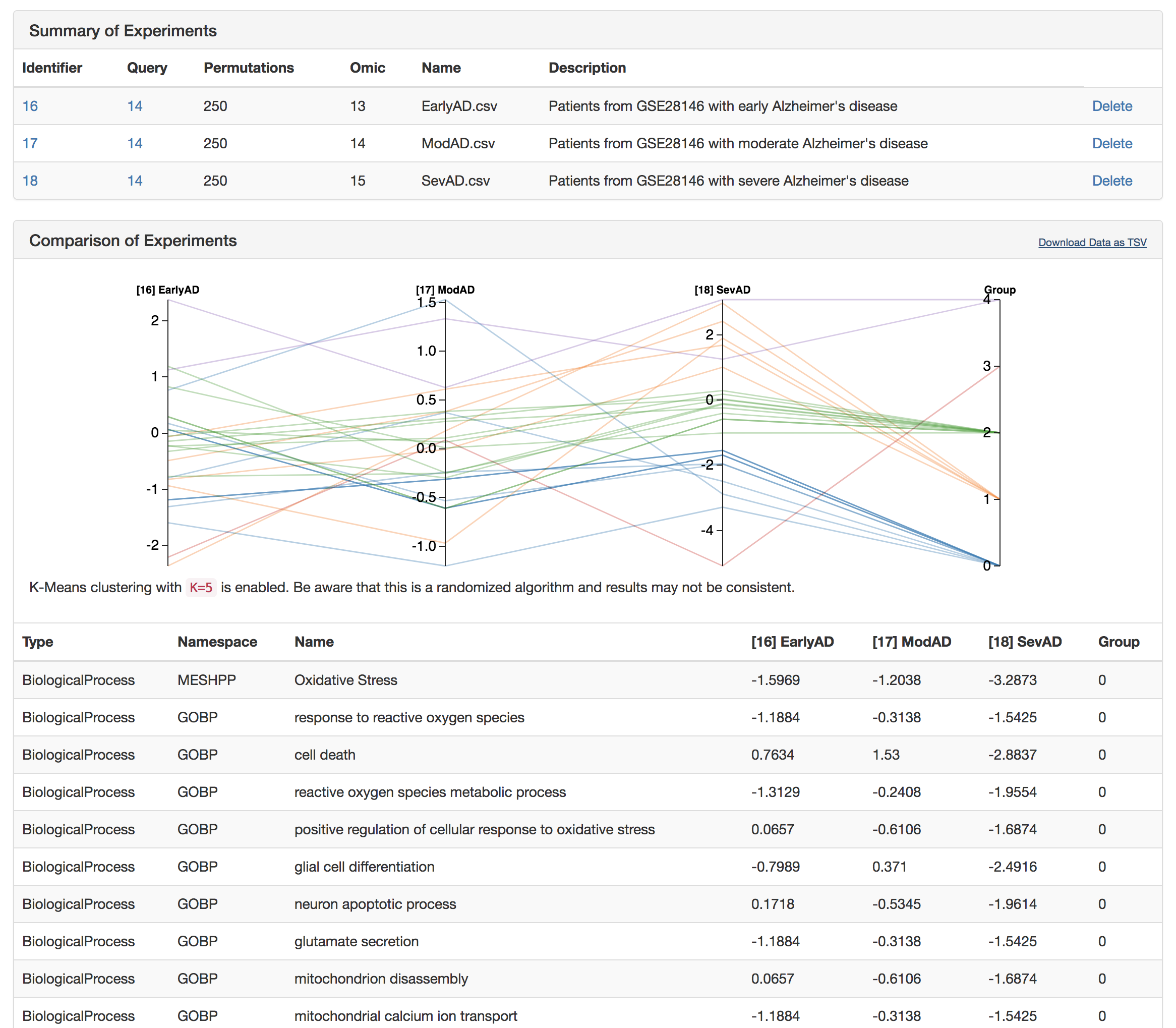
Scroll to "My Experiments" and click "View Comparison"
Interpret the experiments (k-means clustering can be added by appending ?clusters=5 to the end of the URL)