modelsummary creates tables and plots to summarize statistical models and data in R.
The tables produced by modelsummary are beautiful and highly customizable. They can be echoed to the R console or displayed in the RStudio Viewer. They can be saved to a wide variety of formats, including HTML, PDF, Text/Markdown, LaTeX, MS Word, RTF, JPG, and PNG. Tables can easily be embedded in dynamic documents with Rmarkdown, knitr, or Sweave. modelsummary supports dozens of model types out-of-the-box.
Extensive documentation and examples can be found on the modelsummary website: https://vincentarelbundock.github.io/modelsummary
modelsummary includes three families of functions:
modelsummary: Display results from several statistical models side-by-side.modelplot: Plot model coefficients and confidence intervals.datasummary: A flexible tool to create crosstabs and data summaries.datasummary_balance: Balance tables with subgroup statistics and difference in means (aka "Table 1").datasummary_correlation: Correlation tables.datasummary_skim: Quick overview of a dataset.datasummary_df: Turn dataframes into nice tables with titles, notes, etc.
Click on the links at the top of this page to see how these functions are used: https://vincentarelbundock.github.io/modelsummary
These tables and plots were created using modelsummary, without any manual editing at all:
 |
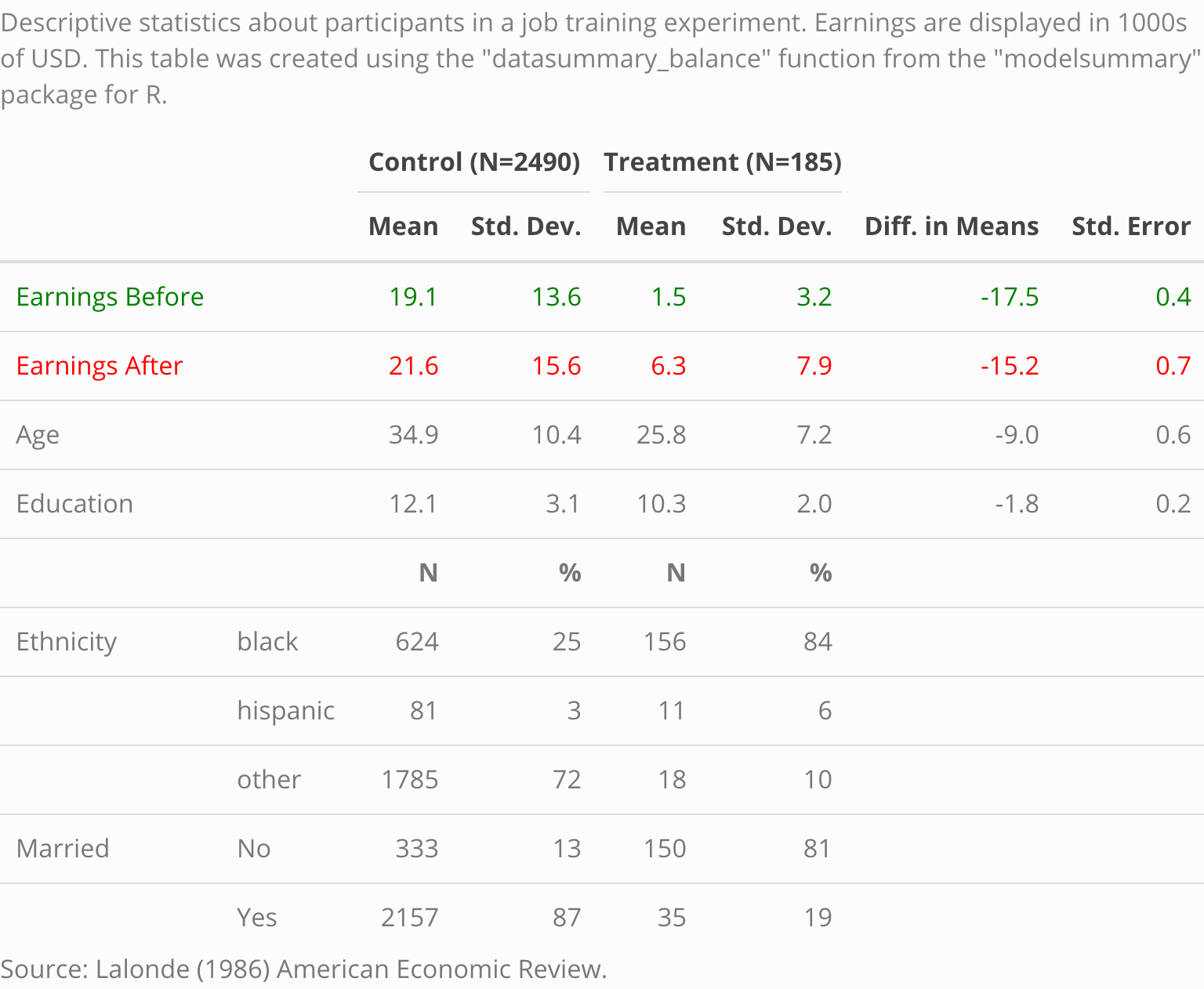 |
 |
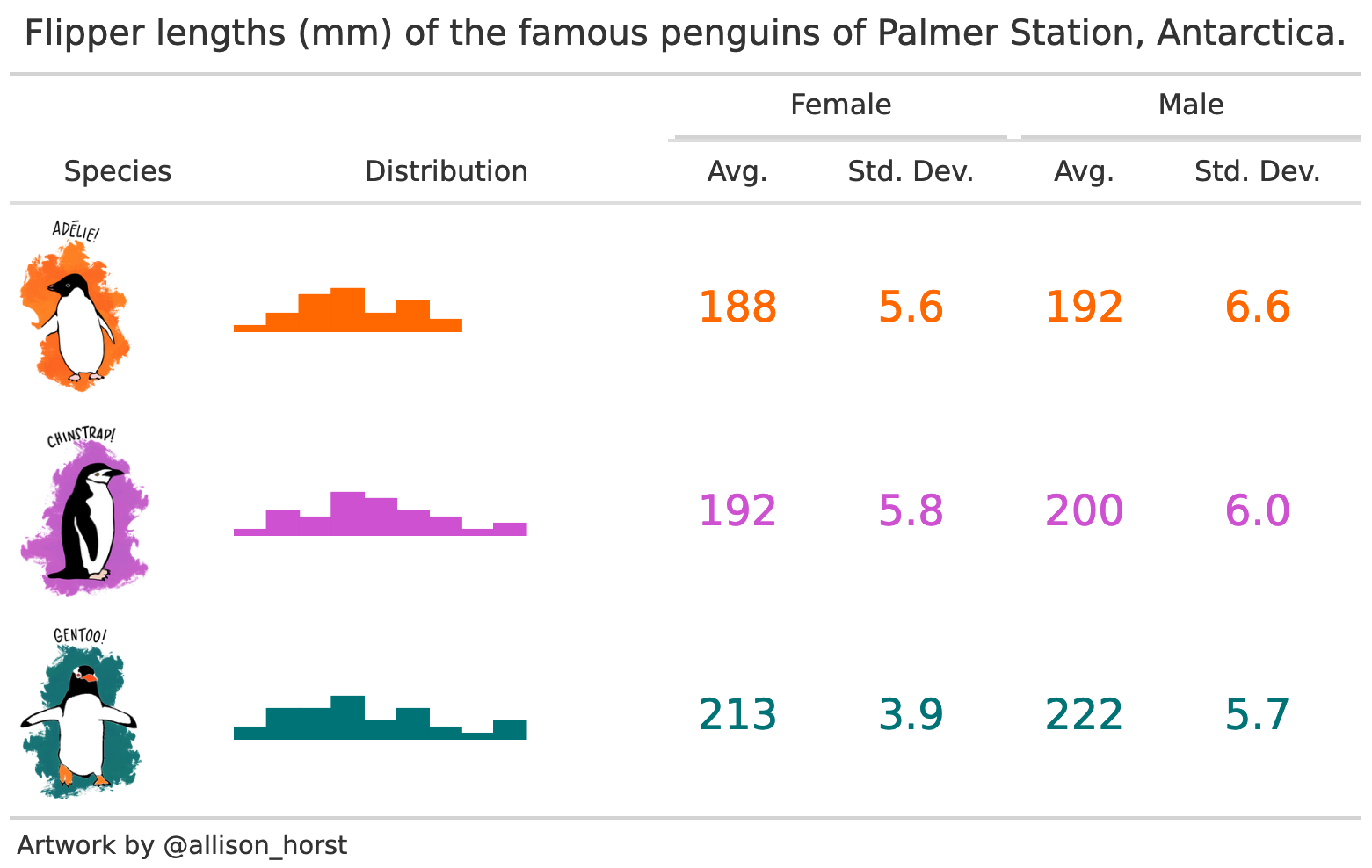 |
 |
 |
 |
- Why should I use
modelsummary? - Installation
- Getting started
- Saving and viewing: output formats
- modelsummary
- modelplot
- datasummary
- Dynamic documents with
Rmarkdownandknitr - Adding and customizing models
- Multiple imputation
- Raw data
Here are a few benefits of modelsummary over some alternative packages:
modelsummary is very easy to use. This simple call often suffices:
library(modelsummary)
mod <- lm(y ~ x, dat)
modelsummary(mod)The command above will automatically display a summary table in the Rstudio Viewer or in a web browser. All you need is one word to change the output format. For example, a text-only version of the table can be printed to the Console by typing:
modelsummary(mod, "markdown")Tables in Microsoft Word and LaTeX formats can be saved to file by typing:
modelsummary(mod, "table.docx")
modelsummary(mod, "table.tex")Information: The package offers many intuitive and powerful utilities to customize the information reported in a summary table. You can rename, reorder, subset or omit parameter estimates; choose the set of goodness-of-fit statistics to include; display various “robust” standard errors or confidence intervals; add titles, footnotes, or source notes; insert stars or custom characters to indicate levels of statistical significance; or add rows with supplemental information about your models.
Appearance: Thanks to the gt, kableExtra, huxtable, and flextable packages, the appearance of modelsummary tables is endlessly customizable. The appearance customization page shows tables with colored cells, weird text, spanning column labels, row groups, titles, source notes, footnotes, significance stars, and more. This only scratches the surface of possibilities.
Supported models: Thanks to the broom package, modelsummary supports dozens of statistical models out-of-the-box. Installing other packages can extend the capabilities further (e.g., broom.mixed). It is also very easy to add or customize your own models.
Output formats: modelsummary tables can be saved to HTML, LaTeX, Text/Markdown, Microsoft Word, Powerpoint, RTF, JPG, or PNG formats. They can also be inserted seamlessly in Rmarkdown documents to produce automated documents and reports in PDF, HTML, RTF, or Microsoft Word formats.
modelsummary is dangerous! It allows users to do stupid stuff like replacing their intercepts by squirrels.
modelsummary is reliably dangerous! The package is developed using a suite of unit tests, so it (probably) won’t break.
modelsummary does not try to do everything. Instead, it leverages the incredible work of the R community. By building on top of the broom package, modelsummary already supports dozens of model types out-of-the-box. modelsummary also supports four of the most popular table-building and customization packages: gt, kableExtra, huxtable, and flextable. packages. By using those packages, modelsummary allows users to produce beautiful, endlessly customizable tables in a wide variety of formats, including HTML, PDF, LaTeX, Markdown, and MS Word.
One benefit of this community-focused approach is that when external packages improve, modelsummary improves as well. Another benefit is that leveraging external packages allows modelsummary to have a massively simplified codebase (relative to other similar packages). This should improve long term code maintainability, and allow contributors to participate through GitHub.
You can install modelsummary from CRAN:
install.packages('modelsummary')If you want the very latest version, install it from Github:
library(remotes)
remotes::install_github('vincentarelbundock/modelsummary')We begin by loading the modelsummary package and by downloading data
from the RDatasets
repository:
library(modelsummary)
url <- 'https://vincentarelbundock.github.io/Rdatasets/csv/HistData/Guerry.csv'
dat <- read.csv(url) We estimate a linear model and call the modelsummary function to display
the results:
mod <- lm(Donations ~ Crime_prop, data = dat)
modelsummary(mod)To summarize multiple models side-by-side, we store them in a list. If the items in that list are named, the names will be used as column labels:
models <- list(
"OLS 1" = lm(Donations ~ Literacy + Clergy, data = dat),
"Poisson 1" = glm(Donations ~ Literacy + Commerce, family = poisson, data = dat),
"OLS 2" = lm(Crime_pers ~ Literacy + Clergy, data = dat),
"Poisson 2" = glm(Crime_pers ~ Literacy + Commerce, family = poisson, data = dat),
"OLS 3" = lm(Crime_prop ~ Literacy + Clergy, data = dat)
)
modelsummary(models)In Rstudio, the image below will be displayed automatically in the
“Viewer” window. When running R from a terminal or from the basic
R interface, this table should appear in your browser.
The same table can be printed in text-only format to the R Console:
modelsummary(models, 'markdown')
| |OLS 1 |Poisson 1 |OLS 2 |Poisson 2 |OLS 3 |
|:-----------|:----------|:-----------|:----------|:-----------|:----------|
|(Intercept) |7948.667 |8.241 |16259.384 |9.876 |11243.544 |
| |(2078.276) |(0.006) |(2611.140) |(0.003) |(1011.240) |
|Clergy |15.257 | |77.148 | |-16.376 |
| |(25.735) | |(32.334) | |(12.522) |
|Literacy |-39.121 |0.003 |3.680 |-0.000 |-68.507 |
| |(37.052) |(0.000) |(46.552) |(0.000) |(18.029) |
|Commerce | |0.011 | |0.001 | |
| | |(0.000) | |(0.000) | |
|Num.Obs. |86 |86 |86 |86 |86 |
|R2 |0.020 | |0.065 | |0.152 |
|Adj.R2 |-0.003 | |0.043 | |0.132 |
|AIC |1740.8 |274160.8 |1780.0 |257564.4 |1616.9 |
|BIC |1750.6 |274168.2 |1789.9 |257571.7 |1626.7 |
|Log.Lik. |-866.392 |-137077.401 |-886.021 |-128779.186 |-804.441 |There are four ways to display and save modelsummary tables.
- Display in the R Console, the RStudio Viewer, or a web browser.
- Save a table to file.
- Insert a table in
Rmarkdownorknitrdocuments, - Convert the table to human-readable html, latex, or markdown code.
To display, simply choose the output format. For example,
modelsummary(models, output = 'latex')
modelsummary(models, output = 'markdown')
modelsummary(models, output = 'gt')
modelsummary(models, output = 'kableExtra')
modelsummary(models, output = 'flextable')
To save a table, choose the file path with the extension you want. For example,
modelsummary(models, output = 'table.tex')
modelsummary(models, output = 'table.docx')
modelsummary(models, output = 'table.html')
modelsummary(models, output = 'table.md')
To customize a table with the gt, kableExtra, flextable, or huxtable packages, choose the output format. Then, you can use functions from those packages to modify the resulting objects:
library(kableExtra)
tab <- modelsummary(models, output = 'kableExtra')
tab %>% row_spec(3, bold = TRUE, color = 'green')
The table-making backends supported by modelsummary have overlaping capabilities (e.g., several of them can produce HTML tables). These are the default packages used for different outputs:
kableExtra:
- HTML
- LaTeX / PDF
flextable:
- Word
- Powerpoint
gt:
- jpg
- png
You can modify these defaults by setting global options such as:
options(modelsummary_html = 'kableExtra')
options(modelsummary_latex = 'gt')
options(modelsummary_word = 'huxtable')
options(modelsummary_png = 'gt')There are several excellent alternatives to draw model summary tables in R:



