Rapid and precise prediction of Mycobacterium tuberculosis complex (MTBC) spoligotype families
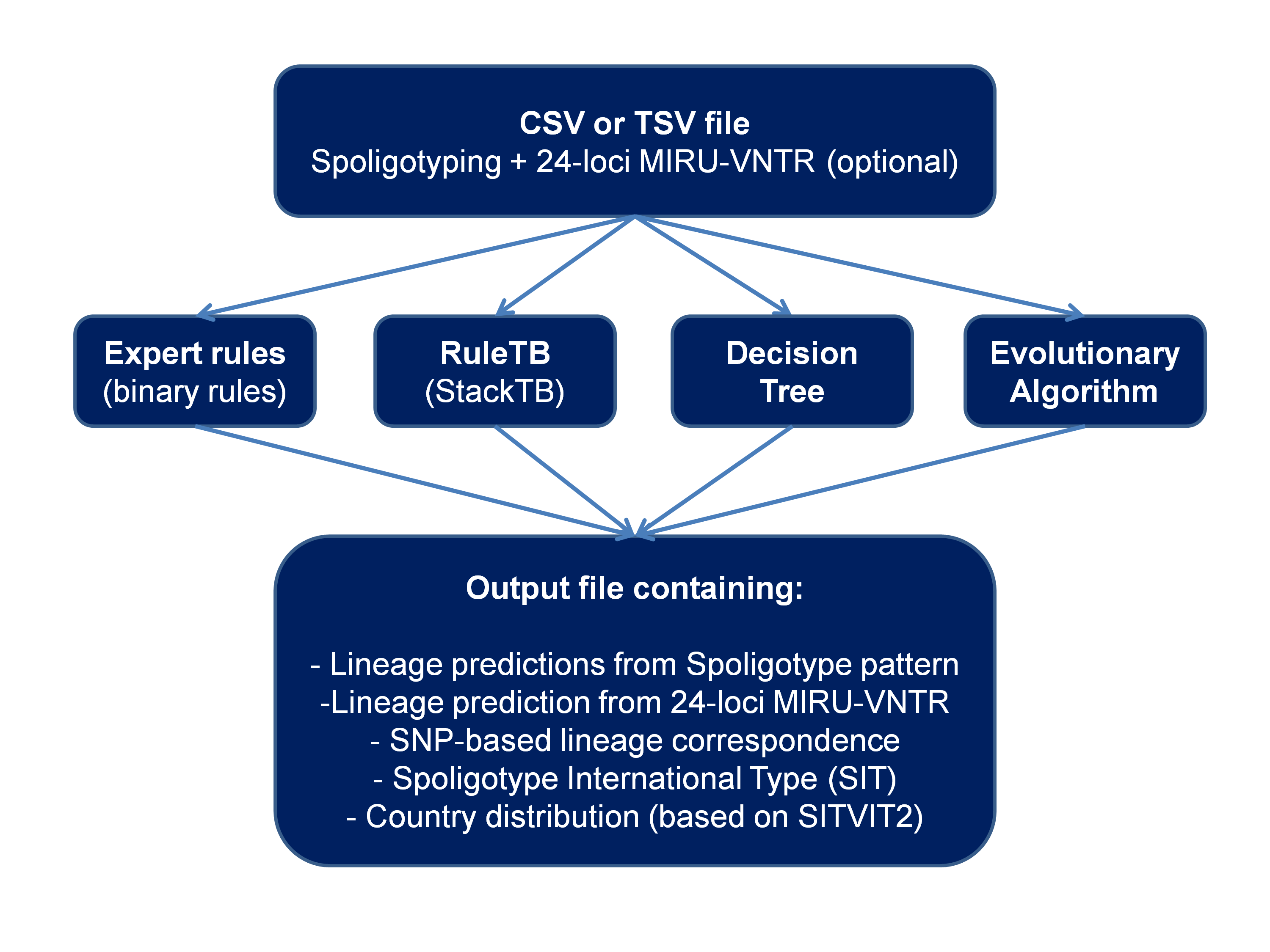
SpolLineages is a software tool mainly written in Java allowing to predict Mycobacterium tuberculosis complex families from spoligotyping or MIRU-VNTR typing patterns using various methods (SITVIT2 binary rules, RuleTB refined rules, Decision tree and/or Evolutionary algorithm).
Java version 6 (or later) programming language must be available in your system to run SpolLineages. Otherwise, you can use the corresponding online tool. Further instructions on how to install Java are available here. To check if Java is installed on your machine, you can run the following command:
java -versionUsers can use the provided executable JAR file and run the following commands for:
-help on this software
java -jar spollineages.jar -h-version
java -jar spollineages.jar -v-before running the evolutionary algorithm (binary mask) prediction, the following commands must be typed (when using Windows, users can install MinGW or Cygwin in order to use the gcc command):
cd Binary_Mask2(when using Unix)
gcc Mask2.c -o Mask2(when using Windows)
gcc Mask2.c -o Mask2.exeor
cd C:\MINGW\bin
gcc.exe path\to\Binary_Mask2\Mask2.c -o path\to\Binary_Mask2\Mask2.exe-Then the Binary Mask folder and Mask2 file should be exported to $PATH (for Unix users) as follows:
export PATH="/path/to/SpolLineages/Binary_Mask2/Mask2:$PATH"
export PATH="/path/to/SpolLineages/Binary_Mask2:$PATH"-run a simple analysis with the provided example (CSV file separated by semicolons)
java -jar spollineages.jar -i example.csv -o result_example.csv-run an example using decision tree or evolutionary algorithm (binary mask) predictions:
java -jar spollineages.jar -i example2.csv -o result_DT.csv -D -pDT C:/Users/dcouvin/workspace/SpolLineages/Decision_Tree/
java -jar spollineages.jar -i example2.csv -o result_EA.csv -E -pEA C:/Users/dcouvin/workspace/SpolLineages/Binary_Mask2/-run an example using both decision tree and evolutionary algorithm predictions as well as all other options:
java -jar spollineages.jar -i example2.csv -o result_DT.csv -D -pDT C:/Users/dcouvin/workspace/SpolLineages/Decision_Tree/ -E -pEA C:/Users/dcouvin/workspace/SpolLineages/Binary_Mask2/ -aIf you use SpolLineages in your work, please cite:
Couvin D, Segretier W, Stattner E, Rastogi N. Novel methods included in SpolLineages tool for fast and precise prediction of Mycobacterium tuberculosis complex spoligotype families. Database (Oxford). 2020 Dec 15;2020:baaa108. https://doi.org/10.1093/database/baaa108 PMID: 33320180