Take CARE of your data!
FAIRly!
- What is CARE-SM for
- How is CARE-SM born?
- What design principles underlie CARE-SM?
- List of defined data elements
- Data model implementation
- Full Documentation
- Communication and feedback
- Cite us
- Acknowledgement
Clinical And Registry Entries (CARE-SM) is a semantic data model designed to effectively represent healthcare patient information by using knowledge graphs represented in the Resource Description Framework (RDF). This technical description aims to provide a comprehensive overview of CARE-SM, its origins, and the design principles that underlie its structure.
CARE-SM is a more robust and matured representation of its precursor, the Common Data Element (CDE) semantic data model. The primary objective of its creation was to develop a semantic data model capable of representing a set of common data elements for rare diseases registration recommended by the European Commission Joint Research Centre. CARE-SM stands as the matured iteration of this CDE semantic model, extending its capabilities to encompass the representation of all data elements pertinent to patient registries and clinical encounters.
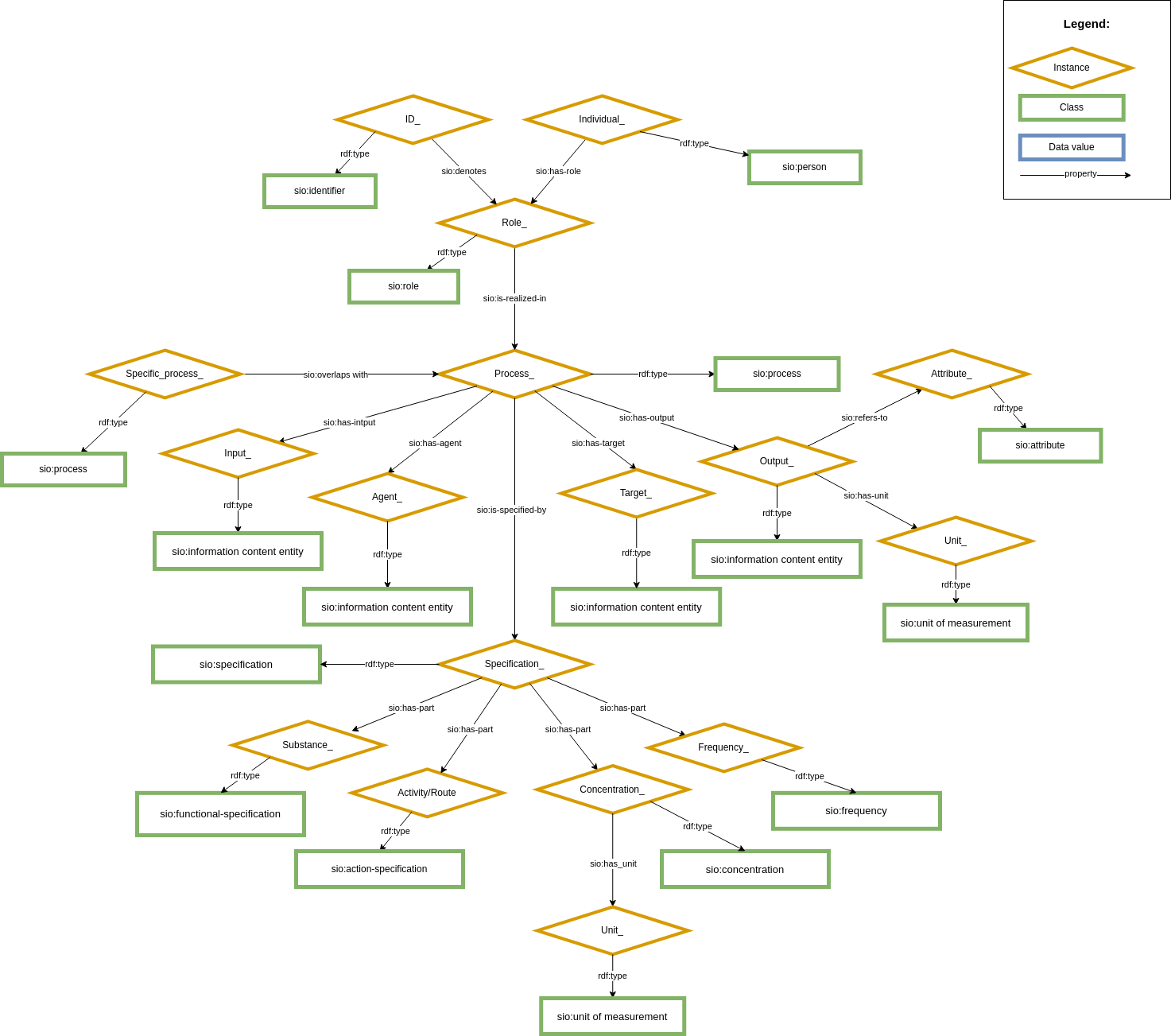
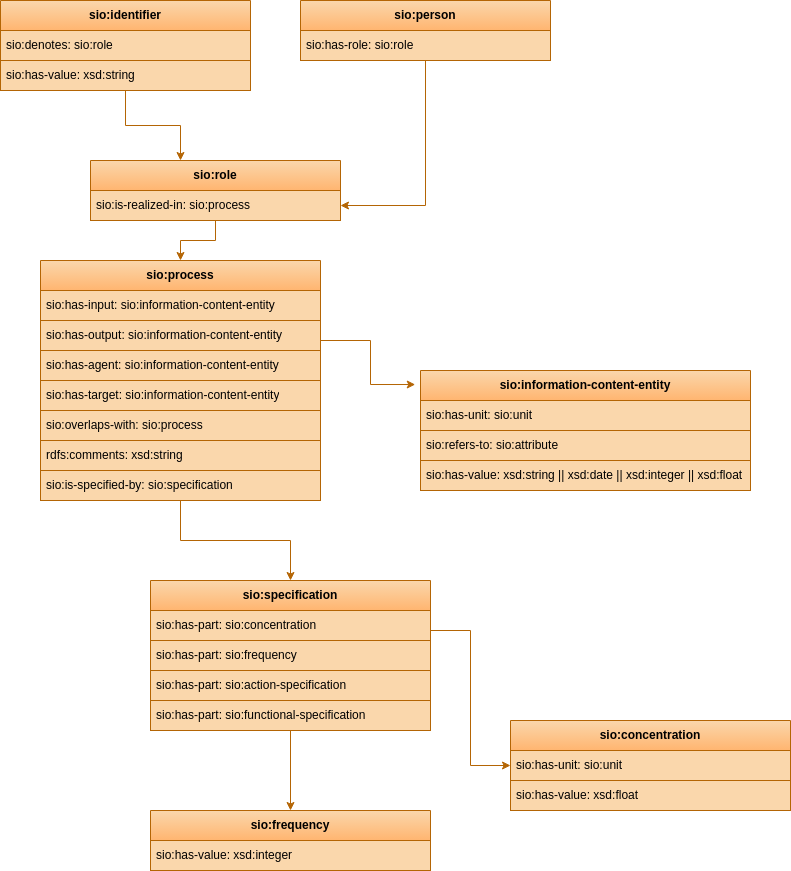
CARE-SM is built upon the Semanticscience Integrated Ontology (SIO) as its core structural schema. SIO is used to define every concept within the data model, utilizing upper-class classes and properties. This knowledge graph serves as an "scaffold" that hold every data element within its structure Figure 1. By a combination of these instances defined by SIO, it becomes possible to represent every clinical entry comprehensively.
Moreover, each instance within CARE-SM is associated with a domain-specific ontological class from the OBO Foundry. For instance, the representation of patient birthdate is described at a upper-class level using the ontological term SIO:attribute and, at a domain-specific level, as NCIT:Birthdate. This dual ontological characterization enhances data interoperability and precise semantic descriptions.
Figure 1: Core structure
Figure 2: Core structure schema
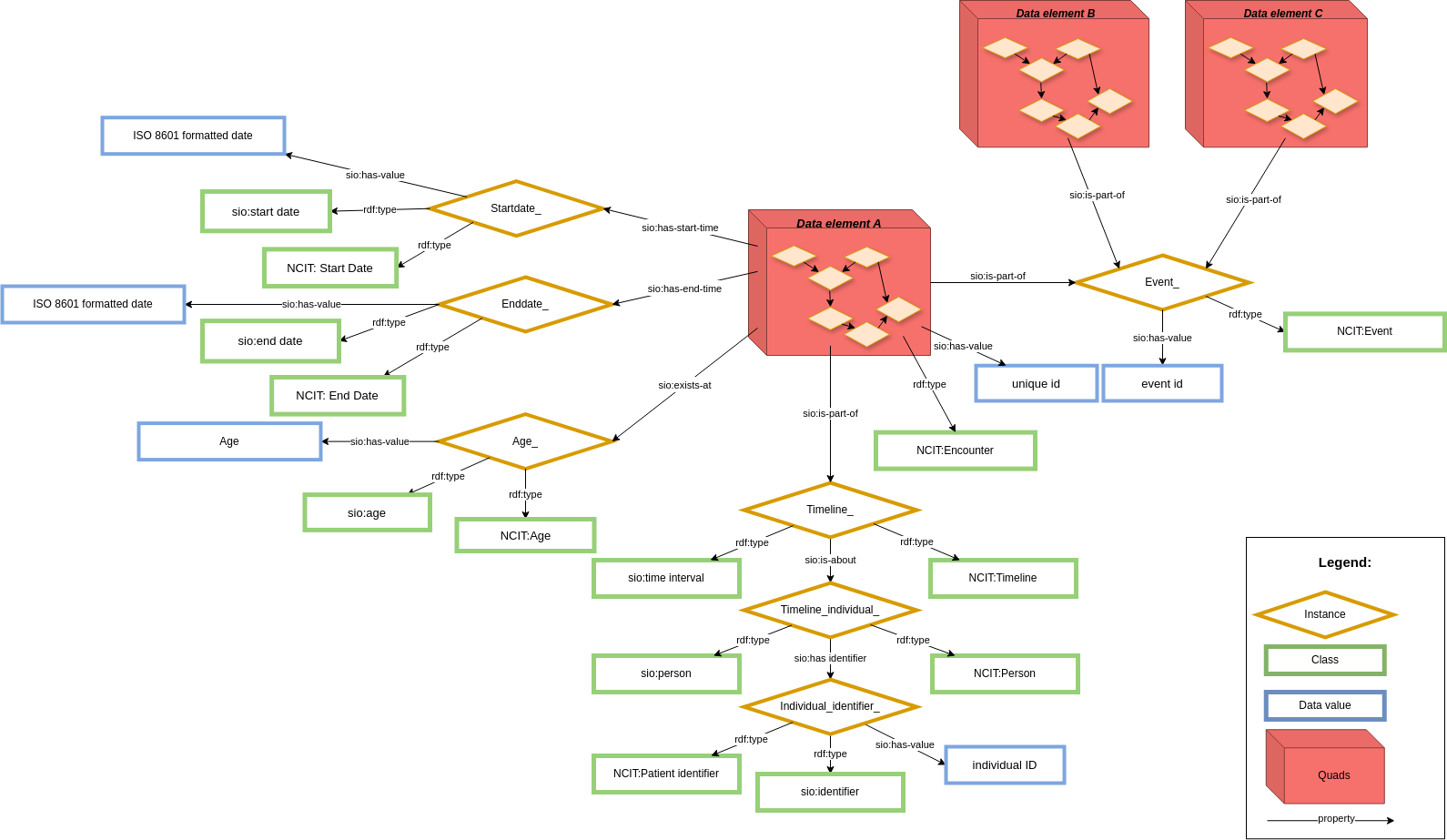
In order to keep a common core structure using CARE-SM, only one data element is modeled at a time. For that reason, if you do not have that element, you do not use that particular data representation. This case could lead to situations where data is not aggregated enough. In order to maintain data aggregated when it's necessary, one layer of metadata has been created around every data element representation (Figure 3).
Its metadata describes the context of the data represented in the core structure model, giving some temporal information to each data element. This structure is kept even when date/time are the core observation of the model (e.g. date of symptom onset). The context layer creates a timeline of events around every data element. Using this timeline, the model is capable of representing not only individual patient registry entries, but also patient clinical encounters in a precise way.
In addition to the patient's timeline and temporal information, common context can be grouped into other arbitrary data elements by connecting them through event nodes. This event has a common context between data elements, for those cases where more than one data element shares a unique relationship (like conditions/treatment scenarios, visit-based aggregated information). It's not mandatory to implement this in your model, it is merely made possible by this model.
This metadata requires the combination of RDF-Quads and RDF-Triples, rather than only RDF Triple used for regular knowledge graphs. The core structure of the model is represented using RDF-Quad, containing as a fourth element (Quad) the same context ID URL. This URL is used as subject for other RDF Triples that define the metadata layer (Figure 3).
Figure 3: Context representation
Based on CARE-SM Core structure, several data element representations can be performed by defining a combination of data model instances, domain-specific ontological terms and its data value. This is a list of data elements presented at patient data registries that can be represented using this data model:
-
Medical history and participation status:
- Birthdate - Patient date of birth.
- Birthyear - Year in which a person was born.
- Deathdate - Patient date of death.
- First confirmed visit - Patient first contact with specialized center.
- Participation status - Patient healthcare participation status.
- Symptoms onset - Patient signs/symptoms onset.
-
Demographic and questionnaire/PROMs representations:
- Sex - Patient sex at birth.
- Education level - Patient scholar level code measured by ISCED.
- Disability - Patient disability score/assessment.
- Questionnaire - Generic questionnaire representation for any clinical question/PROM.
-
Conditions and findings assesments:
- Diagnosis - Patient disease diagnosis.
- Symptom/phenotype assessment - Patient symptom/phenotype assessment.
-
Clinical and molecular measurements:
- Laboratory measurement - Patient laboratory measurements.
- Body measurement - Patient physical measurement of the body.
- Medical imaging - Patient medical imaging data.
- Genetic assessment - Genetic variant assessment.
-
Treatment-related assesments:
- Medication - Patient drug administration based on a prescription.
- Surgical intervention - Therapeutical interventation related to a surgerical procedure.
-
Research sample availability and consent:
-
Clinical trials:
- Clinical trial - patient participation in clinical trial.
This repository primarily focuses on presenting the principles behind the data model and its behavior through a theoretical description.
For information on various workflows for implementing this data model, visit our CARE-SM implementation repository.
See the Wiki for full documentation, examples, operational details and other information.
Your feedback is more than welcome. It will help us improve our semantic data model. Please use github issues to provide your feedback.
To cite this model please use this publication Semantic modeling of common data elements for rare disease registries, and a prototype workflow for their deployment over registry data.
This work was done in the European Joint Programme on Rare Diseases (EJP RD) project which has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement N°82557.