-
Notifications
You must be signed in to change notification settings - Fork 45
Using FABM from a physical model
Note: this is a simple example for a 1D water column model. FABM is built separately as a library. If you would like more detailed information or run into issues, take a look at the documentation further down.
First, create the file <FABMDIR>/src/drivers/<HOSTNAME>/fabm_driver.h (<FABMDIR> is the root of the FABM source tree; <HOSTNAME> the name of your physical model as it will be known to FABM) with the following contents:
#define _FABM_DIMENSION_COUNT_ 1
#define _FABM_DEPTH_DIMENSION_INDEX_ 1
#define _FABM_VECTORIZED_DIMENSION_INDEX_ 1
#include "fabm.h"Now build FABM by creating a directory to build in (typically outside the source tree), and within that, execute:
cmake <FABMDIR> -DFABM_HOST=<HOSTNAME>
make installThis will build the FABM library and place it at ~/local/fabm/<HOSTNAME>/lib. The corresponding include and module files are placed in ~/local/fabm/<HOSTNAME>/include.
The following code demonstrates how to initialize and use FABM when you only have tracers in the interior domain (no surface- or bottom attached state variables), and you are not interested in resolving surface or bottom fluxes. For more advanced applications, please take a look at the detailed documentation below.
use fabm
class (type_fabm_model), pointer :: model
! Initialize (reads FABM configuration from fabm.yaml)
! After this the number of biogeochemical variables is fixed.
! (access variable metadata in model%interior_state_variables, model%interior_diagnostic_variables)
model => fabm_create_model()
! Provide extents of the spatial domain (number of layers nz for a 1D column)
call model%set_domain(nz)
! At this point (after the call to fabm_create_model), memory should be
! allocated to hold the values of all size(model%interior_state_variables) state variables.
! Where this memory resides and how it is laid out is typically host-specific.
! Below, we assume all state variable values are combined in an array interior_state with
! shape nz,size(model%interior_state_variables).
! Point FABM to your state variable data
do ivar = 1, size(model%interior_state_variables)
call model%link_interior_state_data(ivar, interior_state(:,ivar))
end do
! Point FABM to environmental data, here shown for temperature
! Array temp(1:nz) is assumed to be allocated.
! Do this for all variables on FABM's standard variable list that the model can provide.
! For this list, visit https://fabm.net/standard_variables
call model%link_interior_data(fabm_standard_variables%temperature, temp)
! Complete initialization and check whether FABM has all dependencies fulfilled
! (i.e., whether all required calls to model%link_*_data have been made)
call model%start()
! Initialize the tracers
! This sets the values of arrays sent to model%link_interior_state_data,
! in this case those contained in interior_state.
call model%initialize_interior_state(1, nz)
! At this point, initialization is complete.
! Routines below would typically be called every time step.
! Prepare all fields FABM needs to compute source terms (e.g., light)
call model%prepare_inputs()
! Retrieve tracer source terms (tracer units s-1).
! Array dy(1:nz,1:size(model%interior_state_variables)) is assumed to be allocated.
dy = 0
call model%get_interior_sources(1, nz, dy)
! Retrieve vertical velocities (sinking, floating, active movement) in m s-1.
! Array w(1:nz, 1:size(model%interior_state_variables)) is assumed to be allocated.
call model%get_vertical_movement(1, nz, w)
! Compute any remaining diagnostics
call model%finalize_outputs()
! Here you would time-integrate the advection-diffusion-reaction equations
! of all tracers, combining the transport terms with the biogeochemical source
! terms dy and vertical velocities w. This should result in an updated interior_state.Note: this is a simple example for a 3D model with the depth dimension being the third in spatially explicit arrays (i.e., x,y,z), and no spatial mask. FABM is built separately as a library. If you would like more detailed information or run into issues, please take a look at the documentation further down.
First, create the file <FABMDIR>/src/drivers/<HOSTNAME>/fabm_driver.h (<FABMDIR> is the root of the FABM source tree; <HOSTNAME> the name of your physical model as it will be known to FABM) with the following contents:
#define _FABM_DIMENSION_COUNT_ 3
#define _FABM_DEPTH_DIMENSION_INDEX_ 3
#define _FABM_VECTORIZED_DIMENSION_INDEX_ 1
#include "fabm.h"Now build FABM by creating a directory to build in (typically outside the source tree), and within that, execute:
cmake <FABMDIR> -DFABM_HOST=<HOSTNAME>
make installThis will build the FABM library and place it at ~/local/fabm/<HOSTNAME>/lib. The corresponding include and module files are placed in ~/local/fabm/<HOSTNAME>/include.
The following code demonstrates how to initialize and use FABM when you only have tracers in the interior domain (no surface- or bottom attached state variables) and you are not interested in resolving surface or bottom fluxes. For more advanced applications, please take a look at the detailed documentation below.
use fabm
class (type_fabm_model), pointer :: model
! Initialize (reads FABM configuration from fabm.yaml)
! After this the number of biogeochemical variables is fixed.
! (access variable metadata in model%interior_state_variables, model%interior_diagnostic_variables)
model => fabm_create_model()
! Provide extents of the spatial domain.
call model%set_domain(nx, ny, nz)
! At this point (after the call to fabm_create_model), memory should be
! allocated to hold the values of all size(model%interior_state_variables) state variables.
! Where this memory resides and how it is laid out is typically host-specific.
! Below, we assume all state variable values are combined in an array interior_state with
! shape nx, ny, nz, size(model%interior_state_variables).
! Point FABM to your state variable data
do ivar = 1, size(model%interior_state_variables)
call model%link_interior_state_data(ivar, interior_state(:,:,:,ivar))
end do
! Point FABM to environmental data, here shown for temperature
! Array temp with extents nx,ny,nz is assumed to be allocated.
! Do this for all variables on FABM's standard variable list that the model can provide.
! For this list, visit https://fabm.net/standard_variables
call model%link_interior_data(fabm_standard_variables%temperature, temp)
! Complete initialization and check whether FABM has all dependencies fulfilled
! (i.e., whether all required calls to model%link_*_data have been made)
call model%start()
! Initialize the tracers
! This sets the values of arrays sent to model%link_interior_state_data,
! in this case those in interior_state.
do k = 1, nz
do j = 1, ny
call model%initialize_interior_state(1, nx, j, k)
end do
end do
! At this point, initialization is complete.
! Routines below would typically be called every time step.
! Prepare all fields FABM needs to compute source terms (e.g., light)
call model%prepare_inputs()
! In the below, dy and w local to the j,k point being processed.
! They would therefore need to be processed further within the loop to be included in
! the host's advection-diffusion-reaction treatment.
! Alternatively, the could be declared as (4D) global variables that are built up
! in the loop and processed after.
do k = 1, nz
do j = 1, ny
! Retrieve tracer source terms (tracer units s-1).
! Array dy(1:nx, 1:size(model%interior_state_variables)) is assumed to be allocated.
dy = 0
call model%get_interior_sources(1, nx, j, k, dy)
! Retrieve vertical velocities (sinking, floating, active movement) in m s-1.
! Array w(1:nx,1:size(model%interior_state_variables)) is assumed to be allocated.
call model%get_vertical_movement(1, nx, j, k, w)
end do
end do
! Compute any remaining diagnostics
call model%finalize_outputs()
! Here you would time-integrate the advection-diffusion-reaction equations
! of all tracers, combining the transport terms with the biogeochemical source
! terms dy and vertical velocities w. This should result in an updated interior_state.There are no explicit requirements on the physical model, except for the use of the interfaces documented below. However, in order to use FABM efficiently, data for environmental and biotic variables should be stored in a particular way by the host.
Specifically: At any given point in time, all values (across the full domain or subdomain considered by the host) of any single environmental or biotic variable must be accessible through a single Fortran 90 pointer. For spatial models, all data for a single variable must, thus, be stored in a single array (characteristically, a one-dimensional array for column models, or a three-dimensional array for general circulation models). Ultimately, the variable will be represented by a Fortran 90 array slice, which means that the variable for storing is allowed to use additional dimensions (e.g., for time), which are then set to some index chosen by the host, and ignored by FABM.
Note that this does not require that the values for all variables combined are stored in a single array; individual variables may be stored in different arrays. It also does not require that the values of a single variable are contiguous in memory, even though its values should be combined in a single array: it is for instance possible to use an array of shape (nbiotic,nx,ny,nz) to store biotic variables. However, for optimal performance, it is advisable to store a single variable in a contiguous block of memory, which would be achieved by reordering array dimensions as (nx,ny,nz,nbiotic).
Variable values will be accessed through Fortran 90 pointers. Therefore, the Fortran variables at the side of the host that contain environmental or biotic data must be able to serve as the target of a Fortran 90 pointer. That means that it should either have the target or pointer attribute, or be a member of a derived type with one of these attributes.
The arrays on the side of the host could thus be defined with any of the following:
Three examples for a 3D model:
real, dimension(:,:,:), allocatable, target :: temp, salt
real, dimension(nx,ny,nz), target :: temp, salt
real, dimension(:,:,:), pointer :: temp, saltThree examples for a 1D model:
real, dimension(:), allocatable, target :: temp, salt
real, dimension(nz), target :: temp, salt
real, dimension(:), pointer :: temp, saltThey can also be stored as members of a derived type:
type type_data
real, dimension(:,:,:), allocatable :: temp, salt
end typeIn this case, members of the derived type, temp and salt cannot have the target attribute (though they can have the pointer attribute, which would replace allocatable in the above example). To use the target attribute, it must instead be applied to any instance of type type_data that will be linked to FABM, for instance:
type (type_data), target :: datData arrays may also contain additional, non-spatial dimensions. These should be set to a fixed index when used by FABM. For instance, a model may declare arrays as
real, dimension(nx,ny,nz,3), target :: temp, saltWhen sending data to FABM, the last dimension can be ignored by providing slices such as temp(:,:,:,1) and salt(:,:,:,1). This can be used for models that use an additional dimension for the time step index, e.g., MOM4.
If one or more environmental or biotic variables are not stored in the required manner, it will be upon the coupling layer between FABM and its physical host to create arrays of the required structure for these variables, and make sure that the contained values are current upon each call to FABM. It may be clear that this is feasible for variables that have one or more dimensions fewer than the main grid, such as bottom/surface layers, but it will be (very!) expensive if it is to be done for many variables that exist on the full grid.
The physical host is responsible for maintaining (declaring and allocating) spatially explicit arrays for all state variables exported by FABM, and all external variables required by FABM. In the case of state variables, the host must also handle advection and diffusion (if modelled), as well as temporal integration.
State variable values may change through advection and diffusion (if represented in the host model), and by biogeochemical processes represented by FABM. Changes through advection and diffusion must be handled by the host model, that is, if it represents these processes it is also responsible for applying them to the FABM state variables, typically by invoking specific numerical algorithms. In some cases, it is needed to provide these algorithms with the valid range for values taken by the biogeochemical variables represented by FABM, or their positive-definiteness. This can be obtained from FABM structure describing the model (specifically, members minimum and maximum of the elements in array model%interior_state_variables).
FABM provides the changes to the biogeochemical state variables in the form of total temporal derivatives (sink and source terms). That is, it provides the instantaneous rate of change, rather than updated state variable values at the end of a time step. It is the responsibility of the physical host model to perform the (numerical) temporal integration that produces updated state variable values. This is by design. By providing the temporal derivatives, the host model is free to perform temporal integration in any way it sees fit. That may involve splitting of the advection, diffusion and local sink/source operators, or integrate all of these together in a single operation.
FABM generally depends on external variables such as temperature and light; the exact dependencies depend on the set of biogeochemical models that is active. The host model must maintain spatially-explicit arrays with the values of all external variables required by FABM. These may be variables that are already part of the physical host model (e.g., the array in which it stores the current temperature); if they are not part of the physical host model, the driver between FABM and host must declare and allocate the array, and obtain its values from an external data source.
The host model is also responsible for all output.
The physical host must define a number of preprocessor symbols to provide compile-time information about its spatial domain. This includes for instance the number of spatial dimensions (but not their extent, which is set at runtime). These preprocessor definitions are combined in an “include” file that is specific to the physical host. It must be located at src/drivers/<HOSTNAME>/fabm_driver.h. Here, <HOSTNAME> is the name of the host model, e.g., gotm.
The following table lists the preprocessor variables that can be used in the host’s fabm_driver.h.
| symbol | interpretation |
|---|---|
_FABM_DIMENSION_COUNT_ |
The number of dimensions of fully spatially-explicit arrays, such as used for a pelagic variable. Its value can vary from 0 (0D models) to 3 (3D basins). This symbol must always be defined. |
_FABM_DEPTH_DIMENSION_INDEX_ |
Index of the depth dimension. If defined, its value can vary from 1 to _FABM_DIMENSION_COUNT_. If the model has no depth dimension (e.g., if it is vertically integrated), this symbol should not be defined. |
_FABM_VECTORIZED_DIMENSION_INDEX_ |
Index of the dimension that should be vectorized. This makes all FABM subroutines operate on all values across this dimension. If defined, its value can vary from 1 to _FABM_DIMENSION_COUNT_. If not defined, all FABM procedures operate on a local point in space. |
_FABM_BOTTOM_INDEX_ |
This preprocessor symbol specifies whether the vertical index of bottom points is constant (the default; _FABM_BOTTOM_INDEX_=0), or varying in the horizontal (_FABM_BOTTOM_INDEX_=-1). The latter is used for z-coordinate models. This symbol can only be defined if _FABM_DEPTH_DIMENSION_INDEX_ is defined as well. |
_FABM_VERTICAL_BOTTOM_TO_SURFACE_ |
If this preprocessor symbol is defined, it specifies the vertical dimension is ordered from bottom to surface, that is, the vertical index of bottom points is lower than the vertical index of surface points. If this symbol is not defined, vertical ordering is assumed surface to bottom. This symbol can only be defined if _FABM_DEPTH_DIMENSION_INDEX_ is defined as well. |
_FABM_MASK_TYPE_ |
Data type of the spatial mask. This only needs to be defined if part of the spatial domain of the host model is masked, which implies it should be excluded from all biogeochemical calculations (typically, this is done for grid points on land). The data type should match that used internally by the physical host to specify the mask, to allow this mask to be transferred as is (as a pointer) to FABM. It can be any valid Fortran data type (e.g., logical, integer, real). |
_FABM_UNMASKED_VALUE_ |
Value of the host-provided mask that indicates that the grid point must be included in biogeochemical calculations. Conversely, where the mask has any other value, the corresponding point will be excluded from calculations. This symbol must defined only if _FABM_MASK_TYPE_ is defined as well. |
_FABM_HORIZONTAL_MASK_ |
Specifies that the mask is a horizontal-only field, that is, it lacks a depth dimension. If not defined, the mask is a fully spatially-explicit array: it spans the entire pelagic domain. This symbol is used only if _FABM_MASK_TYPE_ is defined as well. |
_FABM_CONTIGUOUS_ |
Specifies that all arrays passed to FABM will be contiguous in memory. This applies to all arrays holding values of state variables and dependencies. Specifying this symbol may allow the compiler to make further optimizations. However, if the arrays you pass at runtime are not contiguous, this will cause problems that are hard to debug. Therefore, use this option only if you are completely sure all arrays will be contiguous. |
After defining these preprocessor symbols, the driver-specific include file must reference FABM's main include file with
#include "fabm.h" A model that describes a 0D well-mixed box would use
#define _FABM_DIMENSION_COUNT_ 0
#include "fabm.h"A model that describes a 1D water column would use
#define _FABM_DIMENSION_COUNT_ 1
#define _FABM_DEPTH_DIMENSION_INDEX_ 1
#define _FABM_VECTORIZED_DIMENSION_INDEX_ 1
#include "fabm.h"A model that describes a 2D vertically-averaged basin would use
#define _FABM_DIMENSION_COUNT_ 2
#define _FABM_VECTORIZED_DIMENSION_INDEX_ 1
#include "fabm.h"A model that describes a 3D basin and has depth varying with its third spatial dimension would typically use the following
#define _FABM_DIMENSION_COUNT_ 3
#define _FABM_DEPTH_DIMENSION_INDEX_ 3
#define _FABM_VECTORIZED_DIMENSION_INDEX_ 1
#include "fabm.h"Note that the typical value for _FABM_VECTORIZED_DIMENSION_INDEX_ (if provided) is 1 in all cases: the best performance in Fortran is generally obtained by looping over the left-most dimension.
Physical host models handle log messages and errors in different ways. In the simplest case, log messages are written to the console, and errors are handled by writing an error message to the console and stopping the process with the stop statement. This is done by default by FABM. However, some models may want to redirect log messages (e.g., to an open file), or to perform additional clean-up when errors are encountered (e.g., by bringing down a parallel model running on multiple cores or machines gracefully).
By default, FABM prints log and error messages to the console (stdout), and halts execution altogether when an error occurs through use of the stop statement. If other behaviour is desired, the host must provide FABM with an object that can handle log messages and errors. To do so, first create a new derived type, extending type_base_driver declared in module fabm_driver:
type, extends(type_base_driver) :: type_<HOSTNAME>_driver
contains
procedure :: fatal_error => <HOSTNAME>_driver_fatal_error
procedure :: log_message => <HOSTNAME>_driver_log_message
end typeSubsequently, add subroutine <HOSTNAME>_driver_fatal_error and <HOSTNAME>_driver_log_message to the same source file. These subroutines should match the following template:
subroutine <HOSTNAME>_driver_fatal_error(self, location, message)
class (type_<HOSTNAME>_driver), intent(inout) :: self
character(len=*), intent(in) :: location,message
write (*,'(a)') trim(location) // ': ' // trim(message)
stop 1
end subroutine
subroutine <HOSTNAME>_driver_log_message(self, message)
class (type_<HOSTNAME>_driver), intent(inout) :: self
character(len=*), intent(in) :: message
write (*,'(a)') trim(message)
end subroutineIn the above, the write and stop statements would be replaced with custom code for handling log messages and fatal errors. Note that in addition to the driver object, subroutine <HOSTNAME>_driver_log_message takes a single argument: the string to log. Conversely, subroutine <HOSTNAME>_driver_fatal_error takes two arguments in addition to the driver object: the subroutine or function triggering the error (a string), and a string description of the error itself. It should be stressed that subroutine fatal_error may never return, which is typically achieved by using the Fortran stop statement directly or indirectly. This prevents data corruption causes by modules continuing to operate after a fatal error occurred.
To provide FABM with the custom logging and error handling routines, make sure that the driver pointer declared in module fabm_driver points to an instance of your derived type, before any of the FABM API routines is called. Typically, this is achieved by:
allocate(type_<HOSTNAME>_driver::driver)Biogeochemical models may depend on variables that the physical host model cannot provide. In this case, the call to model%start will fail due to missing dependencies. It is often desirable to provide the user with a mechanism to fulfil these missing dependencies by providing additional forcing files, for instance, text files or NetCDF with the (time and/or space-varying) values of the required variable. Most physical models already include routines for reading such time and/or space-varying fields. FABM therefore does not provide its own routines, but allows you to leverage the host's routines for this purpose. The implementation of this functionality is highly host specific, but an example could look like this:
! Loop over additional named forcing fields that the user provides
! Below, the current variable is identified by "name"
type (type_fabm_interior_variable_id) :: interior_id
type (type_fabm_horizontal_variable_id) :: horizontal_id
type (type_fabm_scalar_variable_id) :: scalar_id
interior_id = model%get_interior_variable_id(name)
if (model%is_variable_used(interior_id)) then
! Allocate an array to hold the data for this additional interior forcing variable
call model%link_interior_data(interior_id, data)
else
horizontal_id = model%get_horizontal_variable_id(name)
if (model%is_variable_used(horizontal_id)) then
! Allocate an array to hold the data for this additional horizontal forcing variable
call model%link_horizontal_data(horizontal_id, horizontal_data)
else
scalar_id = model%get_scalar_variable_id(name)
if (model%is_variable_used(scalar_id)) then
! Ensure you have a real to hold the value for this additional scalar forcing variable
call model%link_scalar(scalar_id, value)
else
stop 'variable not found'
end if
end if
end ifTypically, you will want to allow the user to provide any number of such additional forcing variables. That means it is easiest to create a linked list of such variables. A single element of this list could be defined by a type such as:
type type_node
type (type_fabm_interior_variable_id) :: interior_id
type (type_fabm_horizontal_variable_id) :: horizontal_id
type (type_fabm_scalar_variable_id) :: scalar_id
real(rk), allocatable :: data(...)
type (type_node), pointer :: next => null()
end typeThe dimensionality of the data member should match that of an interior variable (e.g., data(:,:,:) in a 3D model). When used for horizontal or scalar variables, it would then typically be allocated with one or more of its dimensions having length 1. You could add additional members to this type to hold host-specific information on how to access and/or update the externally provided field. Such members could include NetCDF file or variable identifiers, or unit numbers of open files.
At this point, the list could be started by declaring its head node as follows:
type (type_node), pointer :: first_node => null()While looping over additional forcing variables, you could create new nodes and prepend them to that list with
type (type_node), pointer :: node
allocate(node)
node%next => first_node
first_node => nodeHere is the above example with use of a linked list:
type (type_node), pointer :: node
allocate(node)
node%next => first_node
first_node => node
node%interior_id = model%get_interior_variable_id(name)
if (model%is_variable_used(node%interior_id)) then
! Allocate node%data to hold the data for this additional interior forcing variable
call model%link_interior_data(node%interior_id, node%data)
else
node%horizontal_id = model%get_horizontal_variable_id(name)
if (model%is_variable_used(node%horizontal_id)) then
! Allocate node%data to hold the data for this additional horizontal forcing variable
call model%link_horizontal_data(node%horizontal_id, node%data)
else
node%scalar_id = model%get_scalar_variable_id(name)
if (model%is_variable_used(node%scalar_id)) then
! Allocate node%data to hold the data for single real (this additional scalar forcing variable)
call model%link_scalar(node%scalar_id, node%data)
else
stop 'variable not found'
end if
end if
end ifIf horizontal or scalar variables have fewer dimensions than interior variables, you will need to allocate node%data for those with one or more dimensions having length 1, and slice out those additional dimensions when providing node%data to link_horizonal_data and link_scalar.
If your host itself uses a build system based on CMake, the preferred solution is to directly include FABM's src/CMakeLists.txt from your own CMakeLists.txt file with commands such as:
find_path(FABM_BASE src/fabm.F90 DOC "Path to FABM source directory.")
set(FABM_FORCED_HOST <HOSTNAME>)
add_subdirectory(${FABM_BASE} fabm)(with <HOSTNAME> replaced with the name of your host model) The first line adds a configurable option to the CMake setup for the top-level FABM source directory, the second line preselects the host model that FABM is to be compiled for, and the third line includes the FABM projects.
These CMake commands will add a "fabm" target (a library), which can be referenced in CMake commands such as target_link_libraries. The FABM directory with modules file (*.mod) is part of the public interface of the "fabm" target (it is set in INTERFACE_INCLUDE_DIRECTORIES). This means that any host target that depends on "fabm" will automatically get access to FABM's module files; there is no need to separately set include directories.
By including FABM directly from the host's CMakeLists.txt, the user needs to have the FABM source code on his system, but he does not need to compile or install FABM separately - that will be handled as part of the host compilation. If the source code of either the host or FABM changes, rerunning "make" alone will appropriately recompile all changed files and their dependencies. This is the preferred solution, but only works if the host uses CMake to begin with.
If the host does not use CMake, FABM can be compiled and installed as a library. It can then be linked to while building the host.
For compilation instructions, see the corresponding section on the wiki.
After compiling and installing the FABM library, the FABM library, its include files (*.h) and Fortran 90 module files (*.mod) will have been installed in a directory on the user's system. By default, this directory is ~/local/fabm/<HOSTNAME> on Linux and Mac OS X, %LOCALAPPDATA%\fabm\<HOSTNAME> on Windows Vista and up, and %APPDATA%\fabm\<HOSTNAME> on Windows XP. Within this top-level directory, the following subdirectories are present:
| Setting | Value |
|---|---|
include |
include directory with *.h and *.mod |
lib |
library directory with FABM library (typically libfabm.a on Linux, fabm.lib on Windows) Note that FABM is by default compiled as a static library, though this can be changed by setting BUILD_SHARED_LIBS=ON in CMake. |
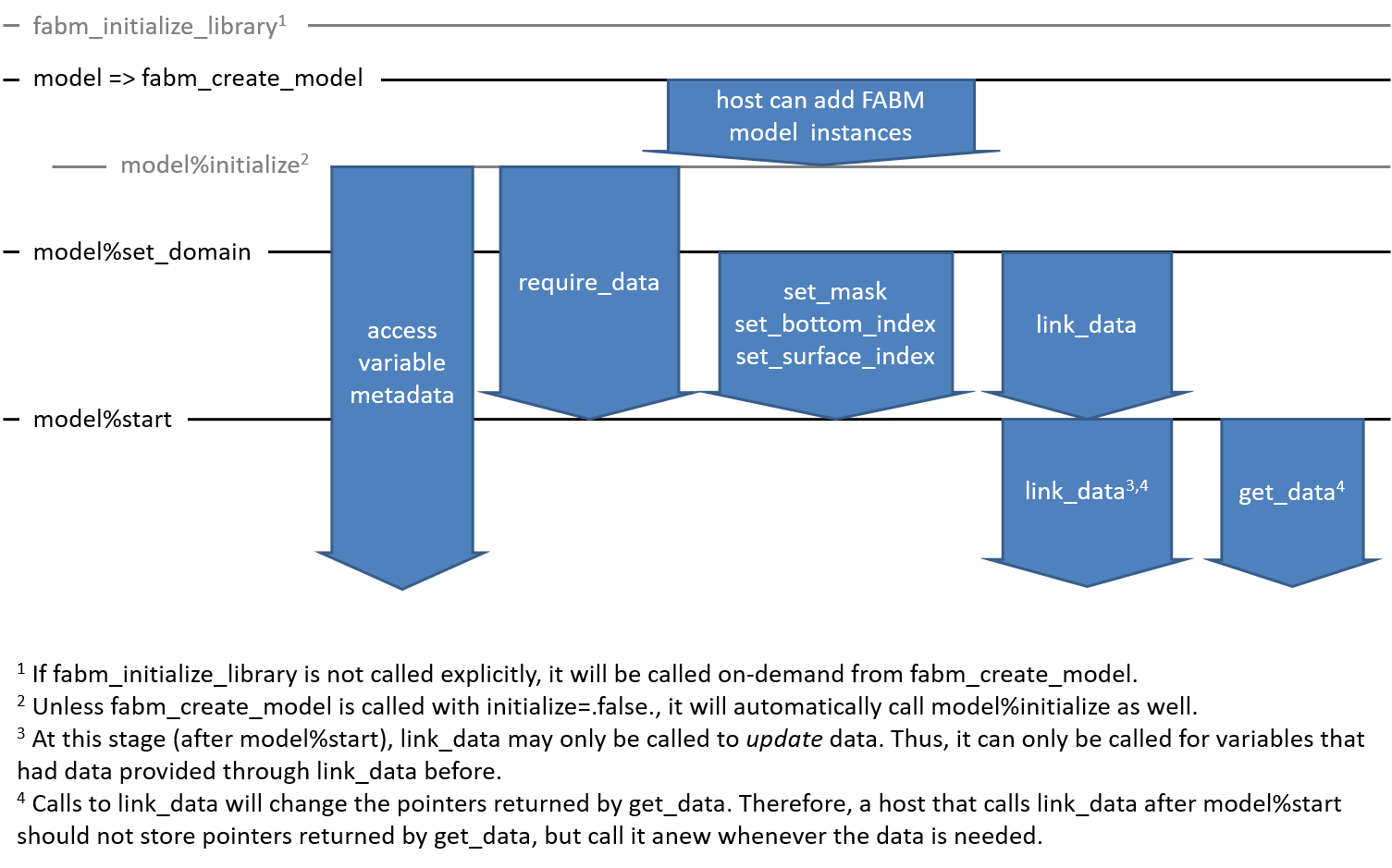
The figure below shows when different routines can or should be called while FABM initializes.
Add a use statement that references the main FABM module, fabm (src/fabm.F90):
use fabmDeclare a single model object, with derived type type_fabm_model:
class (type_fabm_model), pointer :: modelThis instance will hold metadata on all biogeochemical models active within FABM. Metadata include information such as the name and units of state variables, diagnostic variables, conserved quantities and model parameters.
If you use you own routines for logging and error handling, you need to provide these before calling FABM. To do so, allocate FABM's driver as an object of your own error handling type:
allocate(type_<HOSTNAME>_driver::driver)Note that the driver variable is declared in module fabm_driver.
Initialize the model by calling fabm_create_model:
model => fabm_create_model()This routine reads FABM's run-time configuration and stores it in the model object. After this call, the model structure is frozen, which means you can enumerate all model variables and parameters. You can still change the selection of diagnostics that FABM will compute and store, by modifying the attribute save (logical) of elements of model%interior_diagnostic_variables and model%horizontal_diagnostic_variables.
fabm_create_model takes several optional arguments:
-
file(a string). This argument specifies the path to the file from which the FABM configuration must be read (default:fabm.yamlin the current working directory). It is strongly recommended to use this default path, as users will expect this, but in special cases (e.g., a need to switch between different configuration files through command line arguments) you may want to explicitly set the path. -
initialize(a logical). This argument specifies whether to initialize the models (as well as create them). This is.true.by default. Set it to.false.if you programmatically want to add additional FABM instances, not specified in the FABM configuration file (fabm.yaml). In that case, you will have to callmodel%initializemanually after all model instances have been added. -
unit(an integer). If provided, the YAML configuration will be read directly from this unit; no file will be opened or closed. This is useful if host-specific routines for opening and closing the file need to be used.
Provide FABM with the extents of the spatial domain:
call model%set_domain(<DOMAINSIZE>)Here, <DOMAINSIZE> is a placeholder for a comma-separated set of 0, 1 or more integers, one per spatial dimension; the number of integers must equal the value of _FABM_DIMENSION_COUNT_. For instance, in the 1D water column model GOTM, <DOMAINSIZE> is nlev, which specifies the number of layers in the water column. For the 3D model MOM4, <DOMAINSIZE> is iec-isc+1,jec-jsc+1,nz, with isc and jsc specifying the start index of the x- and y dimensions of the computational domain, iec and jec specifying the end index of the x and y dimensions of the computational domain, and nz specifying the number of vertical layers.
This is optional: If the host model has a vertical dimension and the index of the bottom layer varies in the horizontal (preprocessor symbols _FABM_DEPTH_DIMENSION_INDEX_ and _FABM_BOTTOM_INDEX_=-1 are defined in fabm_driver.h), you need to tell FABM at which vertical indices it can find the bottom values of interior variables:
call model%set_bottom_index(bottom_indices)Here, bottom_indices is an array of integers, with dimensionality matching that of horizontal fields, and extents matching the values provided to set_domain. As FABM will keep a pointer to this array, the indices are allowed to change during the simulation. This can be useful when vertical layers are dynamically split and merged, as in some isopycnal coordinate models. As FABM keeps a pointer to the array, it must be declared with target or pointer attribute at the side of the host (if the array is a member of a derived type, the target attribute should be applied to the type instance rather than the type member).
This is optional: If the host model masks part of the spatial domain, i.e., if preprocessor symbol _FABM_MASK_TYPE_ is defined, you must provide FABM with (a pointer to) the mask:
call model%set_mask(mask)Argument mask has the data type defined by preprocessor symbol _FABM_MASK_TYPE_. Its number of dimensions should equal those of the spatial domain, i.e., _FABM_DIMENSION_COUNT_, unless preprocessor symbol _FABM_HORIZONTAL_MASK_ is defined, in which case its number of dimensions must match that of horizontal-only fields (it excludes the depth dimension). The extents of the dimensions of the mask should match those provided to set_domain.
The provided mask should be allowed to serve as a pointer target, which implies it must be declared with target or pointer attribute at the side of the host (if the mask is a member of a derived type, the target attribute should be applied to the type instance rather than the type member).
As FABM will keep a pointer to the mask, it is allowed to change during the simulation. Thus, simulations that include flooding and drying can dynamically exclude dry regions from FABM computations by updating the mask.
Provide FABM with pointers to arrays that will hold the current values of host-controlled environmental variables, such as temperature and salinity. Each array must extend across its entire relevant spatial domain (e.g., all of the pelagic). Variables may be defined on the interior model domain (e.g., temperature), or on a horizontal slice only (typically for bottom or surface, e.g., wind speed, bottom stress).
Variables are provided by calling model%link_interior_data, model%link_horizontal_data, and model%link_scalar. These subroutines take two arguments:
- The variable identifier. This must be of type
type_interior_standard_variable,type_horizontal_standard_variable, ortype_global_standard_variablefor interior, horizontal and scalar variables respectively. Many identifiers are predefined as members of the objectfabm_standard_variables. If a suitable identifier does not exist yet, you can create one withtype_interior_standard_variable(name=<NAME>,units=<UNITS>)for variables in the domain interior, andtype_horizontal_standard_variable(name=<NAME>,units=<UNITS>)for variables defined on a horizontal slice of the domain. In both cases,<NAME>is a short variable name (letters and underscores only), and<UNITS>is a unit specifier (both strings). Where possible, it is recommended to follow the CF guidelines for constructing standard names, along with the simplifications defined by FABM near the top ofsrc/fabm_standard_variables.F90. This is the same approach taken by FABM, which means that when in time your custom standard variable is included in FABM's own list, the chance is high that its name will be the same, allowing code based on your custom standard variable to keep working. - The array slice or scalar that will hold the data. This variable must be declared with the
targetorpointerattribute by the host model, to allow FABM to keep a pointer to it. It must be of typereal(rk). Its dimensionality must match the full extents of the relevant spatial domain, as provided toset_domain.
For instance, a 3D model might declare arrays with temperature and practical salinity as
real, dimension(nx,ny,nz), target :: temp, saltThese should be provided to FABM as follows:
call model%link_interior_data(fabm_standard_variables%temperature, temp)
call model%link_interior_data(fabm_standard_variables%practical_salinity, salt)Here, model is the model object declared and initialized above, and temperature and practical_salinity are members of the fabm_standard_variables object provided by the fabm module. A list of all standard variables predefined by FABM is maintained here.
Similarly, variables defined on a horizontal slice of the domain are provided by calling subroutine model%link_horizontal_data. For instance, a 3D model might have an array with surface wind speed values allocated as
real, dimension(nx,ny), target :: windThis should be provided to FABM as follows:
call model%link_horizontal_data(fabm_standard_variables%wind_speed, wind)Finally, variables that are independent of the spatial domain are provided by calling subroutine model%link_scalar. A model that declares a variable that will hold the day of the year as
real,target :: yeardayshould provide it to FABM by calling
call model%link_scalar(fabm_standard_variables%number_of_days_since_start_of_the_year, yearday)Subroutines link_interior_data, link_horizontal_data and link_scalar may be called at any time during program execution, and they may be called repeatedly with the same variable identifier. Thus, it is possible to redirect FABM to another in-memory array slice during simulation. If this is desired, however, it is for performance reasons strongly recommended that you first obtain a runtime variable identifier, once, at the start of the simulation, by calling model%get_interior_variable_id, model%get_horizontal_variable_id or model%get_scalar_variable_id. This identifier can subsequently be substituted for the standard variable object in calls to link_interior_data, link_horizontal_data and link_scalar. For instance,
type (type_fabm_interior_variable_id) :: id_temp
id_temp = model%get_interior_variable_id(fabm_standard_variables%temperature)
call model%link_interior_data(id_temp, temp)
type (type_fabm_horizontal_variable_id) :: id_wind
id_wind = model%get_horizontal_variable_id(fabm_standard_variables%wind_speed)
call model%link_horizontal_data(id_wind, wind)
type (type_fabm_scalar_variable_id) :: id_yearday
id_yearday = model%get_scalar_variable_id(fabm_standard_variables%number_of_days_since_start_of_the_year)
call model%link_scalar(id_yearday, yearday)In some cases you may be able to compute certain environmental variables from the quantities described by the physical model (e.g., pressure from layer heights, density and gravity), but as that computation is computationally expensive or memory-consuming, you would want to do so only if that environmental variable is actually required by the current selection of biogeochemical models. This is possible. You can call model%variable_needs_values to determine whether a particular variable is required by the current selection of biogeochemical models, e.g.,
if (model%variable_needs_values(fabm_standard_variables%pressure)) then
allocate(pres(nx, ny, nz)
call model%link_interior_data(fabm_standard_variables%pressure, pres)
end ifAt the start of each time step you would then update the pressure values, before calling FABM. For instance:
if (allocated(pres)) then
! Compute pressure from layer heights, density and gravity.
end ifCreate spatially-explicit arrays that will hold the values for the state variables exported by FABM. These typically include state variables defined on the full (3D pelagic) model domain, but they may also include bottom-bound (benthic) and surface-bound variables, which are defined on a horizontal slice of the domain only.
A 3D model could define and allocate the following arrays to store state variable values:
real, dimension(:,:,:,:), target, allocatable :: state
real, dimension(:,:,:) , target, allocatable :: bottom_state, surface_state
allocate(state (nx, ny, nz, size(model%interior_state_variables))
allocate(bottom_state (nx, ny, size(model%bottom_state_variables))
allocate(surface_state(nx, ny, size(model%surface_state_variables))A 1D water column model would not include the x and y dimensions, but otherwise could do it the same way.
After defining and allocating the arrays, they must linked to FABM by calling subroutines model%link_interior_state_data, model%link_bottom_state_data and model%link_surface_state_data, for pelagic, bottom-bound and surface-bound state variables, respectively. Data must be provided individually for each variable. If arrays are defined as in the above example, these could be transferred as follows:
do ivar=1,size(model%interior_state_variables)
call model%link_interior_state_data(ivar, state(:,:,:,ivar))
end do
do ivar=1,size(model%bottom_state_variables)
call model%link_bottom_state_data(ivar, bottom_state(:,:,ivar))
end do
do ivar=1,size(model%surface_state_variables)
call model%link_surface_state_data(ivar, surface_state(:,:,ivar))
end doIt is worth noting that model%link_interior_state_data, model%link_bottom_state_data and model%link_surface_state_data may be called at any time during program execution, and that they may be called repeatedly with the same variable index. Thus, it is possible to redirect FABM to another in-memory array slice during simulation, if needed.
After you have provided the model with all information (domain extent, mask if using, state, environment, etc.), you call model%start to tell FABM that the model is now complete. FABM then verifies whether it has all data it needs (pointers to variable data; the spatial mask if used):
call model%start()FABM will report which data are still missing, if any. In case of missing data, FABM will stop with a fatal error.
After calling start, the selection of diagnostics that FABM will compute and store is frozen; modification of the save attribute of elements of model%interior_diagnostic_variables and model%horizontal_diagnostic_variables no longer has any effect.
Unless you are initializing the model with a state stored previously (e.g., in a restart file), you will want FABM to set initial values for its interior, surface-attached and bottom-attached state variables. These are typically values that are the same across the entire spatial domain, but models can in principle also compute space-dependent initial values from properties of the environment (e.g., from bathymetry values).
To let FABM models set their initial state, call model%initialize_state(<LOCATION>), model%initialize_surface_state(<HORIZONTAL_LOCATION>) and model%initialize_bottom_state(<HORIZONTAL_LOCATION>).
Call model%prepare_inputs() to let FABM compute any fields it needs to later compute source terms and fluxes across the surface or bottom. Commonly, this includes the interior light field, which in turn might require light attenuation or other inputs.
call model%prepare_inputs()This routine operates on the entire active spatial domain and therefore does not take any arguments.
Call model%get_surface_sources(<HORIZONTAL_LOCATION>, flux_sf, sms_sf) to get fluxes of pelagic state variables over the air-water interface (flux_sf) as well as source terms of surface-attached state variables (sms_sf). Both have typically have units matter m-2 s-1: fluxes for pelagic state variables have unit "variable units × m s-1", source terms for surface-attached variables have unit "variable units s-1". For flux_sf, a positive value indicates a flux into the water; a negative value indicates a flux out of the water. Important: FABM increments rather than sets arguments flux_sf and sms_sf; therefore, you will typically want to initialize these arguments to zero before calling get_surface_sources.
The host is responsible for updating the value of surface-attached and pelagic state variables according to the value of the returned source terms and surface fluxes. Source terms for surface-attached state variables are typically processed by directly time-integrating the variables. Surface fluxes for pelagic state variables can be accounted for in different ways: they could be used as flux (Neumann) boundary condition in advection or diffusion solvers, or they could be added to the pelagic source terms after dividing the surface flux by the height of the uppermost layer. For consistency (and mass conservation!), it is preferable to process source terms and surface fluxes with the same numerical scheme, which often implies that surface fluxes are best included among pelagic source terms and then time-integrated.
Some common examples:
! 0D box
real(rk) :: flux_sf(size(model%interior_state_variables))
real(rk) :: sms_sf(size(model%surface_state_variables))
flux_sf = 0
sms_sf = 0
call model%get_surface_sources(flux_sf, sms_sf)
! 1D water column with extents nz
real(rk) :: flux_sf(size(model%interior_state_variables))
real(rk) :: sms_sf(size(model%surface_state_variables))
flux_sf = 0
sms_sf = 0
call model%get_surface_sources(flux_sf, sms_sf)
! 3D basin with extents nx,ny,nz
real(rk) :: flux_sf(nx,size(model%interior_state_variables))
real(rk) :: sms_sf(nx,size(model%surface_state_variables))
do j=1,ny
flux_sf = 0
sms_sf = 0
call model%get_surface_sources(1, nx, j, flux_sf, sms_sf)
! Here you would use or store the returned surface fluxes and source terms
end doCall model%get_bottom_sources(<HORIZONTAL_LOCATION>, flux_bt, sms_bt) to get fluxes of pelagic state variables over the pelagic-benthic interface (flux_bt) as well as source terms of bottom-attached state variables (sms_bt). Both have typically have units matter m-2 s-1: fluxes for pelagic state variables have unit "variable units × m s-1", source terms for bottom-attached variables have unit "variable units s-1". For flux_bt, a positive value indicates a flux into the water; a negative value indicates a flux out of the water. Important: FABM increments rather than sets arguments flux_bt and sms_bt; therefore, you will typically want to initialize these arguments to zero before calling get_bottom_sources.
The host is responsible for updating the value of bottom-attached and pelagic state variables according to the value of the returned source terms and surface fluxes. Source terms for bottom-attached state variables are typically processed by directly time-integrating the variables. Bottom fluxes for pelagic state variables can be accounted for in different ways: they could be used as flux (Neumann) boundary condition in advection or diffusion solvers, or they could be added to the pelagic source terms after dividing the bottom flux by the height of the bottommost layer. For consistency (and mass conservation!), it is preferable to process source terms and bottom fluxes with the same numerical scheme, which often implies that bottom fluxes are best included among pelagic source terms and then time-integrated.
Some common examples:
! 0D box
real(rk) :: flux_bt(size(model%interior_state_variables))
real(rk) :: sms_bt(size(model%bottom_state_variables))
flux_bt = 0
sms_bt = 0
call model%get_bottom_sources(flux_bt, sms_bt)
! 1D water column with extents nz
real(rk) :: flux_bt(size(model%interior_state_variables))
real(rk) :: sms_bt(size(model%bottom_state_variables))
flux_bt = 0
sms_bt = 0
call model%get_bottom_sources(flux_bt, sms_bt)
! 3D basin with extents nx,ny,nz
real(rk) :: flux_bt(nx,size(model%interior_state_variables))
real(rk) :: sms_bt(nx,size(model%bottom_state_variables))
do j=1,ny
flux_bt = 0
sms_bt = 0
call model%get_bottom_sources(1, nx, j, flux_bt, sms_bt)
! Here you would use or store the returned bottom fluxes and source terms
end doCall model%get_interior_sources(<LOCATION>,sms) to get the source terms sms of pelagic state variables, in variable units s-1). Important: FABM increments rather than sets argument sms; therefore, you will typically want to initialize this argument to zero before calling get_interior_sources.
The host is responsible for time-integrating the value of pelagic state variables according to the returned source terms. FABM does not specify how this is to be done: it is conceivable to simultaneously time-integrate source terms and transport (advection/diffusion) terms, but one can also apply operator splitting to separately time-integrate the source terms, while transport is handled with specialized numerical schemes.
Some common examples:
! 0D box
real(rk) :: sms(size(model%interior_state_variables))
sms = 0
call model%get_interior_sources(sms)
! 1D water column with extents nz
real(rk) :: sms(nz,size(model%interior_state_variables))
sms = 0
call model%get_interior_sources(1, nz, sms)
! 3D basin with extents nx,ny,nz
real(rk) :: sms(nx,size(model%interior_state_variables))
do k=1,nz
do j=1,ny
sms = 0
call model%get_interior_sources(1, nx, j, k, sms)
! Here you would use or store the returned source terms
end do
end doCall model%get_vertical_movement(<LOCATION>, velocity) to get local vertical velocities of pelagic state variables. These describe movement through the water, independent of water flow, i.e., sinking, floating or active movement of state variables. On return, velocity contains velocities in m s-1 of all pelagic state variables. Rates are negative for downward movement (sinking, sedimentation), positive for upward movement (floating).
The host must update state variable values according to these movement rates, typically by applying an advection scheme to the vertical movement term.
Some common examples:
! 0D box
real(rk) :: velocity(size(model%interior_state_variables))
call model%get_vertical_movement(velocity)
! 1D water column with extents nz
real(rk) :: velocity(nz,size(model%interior_state_variables))
call model%get_vertical_movement(1, nz, velocity)
! 3D basin with extents nx,ny,nz
! You may want to swap the k and j loops below, in order handle a complete
! i,k slice with an advection scheme before moving to the next j.
real(rk) :: velocity(nx,size(model%interior_state_variables))
do k=1,nz
do j=1,ny
call model%get_vertical_movement(1, nx, j, k, velocity)
! Here you would use or store the returned vertical velocities
end do
end doCall model%finalize_outputs() to let FABM compute any remaining outputs (diagnostics) that were not already computed by preceding routines (get_surface_sources, get_bottom_sources, get_interior_sources, etc.):
call model%finalize_outputs()This routine operates on the entire active spatial domain and therefore does not take any arguments.
Optional: these totals can be used to check mass conservation
Call model%get_interior_conserved_quantities(<LOCATION>, total) to get the value of the conserved quantities described by the model for the interior domain (e.g., the pelagic). This routine returns one value per conserved quantity, in units defined by the biogeochemical models during their initialization phase. Names, long names and units of conserved quantities are available from the model%conserved_quantities array.
Call model%get_horizontal_conserved_quantities(<HORIZONTAL_LOCATION>, total_hz) to get the totals of conserved quantities at the surface and bottom interfaces. This routine returns one value per conserved quantity, in units defined by the biogeochemical models during their initialization phase.
Typically, the host calls these routines over the entire spatial domain, multiplies entries in total by the volume of the associated grid cells, multiplies entries in total_sf with the surface area of the associated grid cells, then sums all values to arrive at a total (a real number) for the entire domain. This number is the included in the output or presented to the user, so it can be used to verify whether the model conserves mass.
Metadata for conserved quantities are available from arrays model%conserved_quantities. Among others, elements of this arrays have members name, long_name, and units (all strings).
Some common examples:
! 0D box
real(rk) :: H ! Height of box in m, assumed to be set already
real(rk) :: total_interior(size(model%conserved_quantities))
real(rk) :: total_interfaces(size(model%conserved_quantities))
real(rk) :: total(size(model%conserved_quantities)) ! Totals in matter per m2!
call model%get_interior_conserved_quantities(total_interior)
call model%get_horizontal_conserved_quantities(total_interfaces)
total = H*total_interior + total_interfaces
! 1D water column with extents nz
real(rk) :: H(nz) ! Height of the layers in m, assumed to be set already
real(rk) :: total_interior(nz,size(model%conserved_quantities))
real(rk) :: total_interfaces(size(model%conserved_quantities))
real(rk) :: total(size(model%conserved_quantities)) ! Totals in matter per m2!
call model%get_interior_conserved_quantities(1, nz, total_interior)
call model%get_horizontal_conserved_quantities(total_interfaces)
do ivar=1,size(model%conserved_quantities)
total(ivar) = sum(H*total_interior(:,ivar)) + total_interfaces(ivar)
end do
! 3D basin with extents nx,ny,nz
real(rk) :: A(nx,ny) ! Surface area of the cells in m2, assumed to be set already
real(rk) :: V(nx,ny,nz) ! Volume of the cells in m3, assumed to be set already
real(rk) :: total_interior(nx,size(model%conserved_quantities))
real(rk) :: total_interfaces(nx,size(model%conserved_quantities))
real(rk) :: total(size(model%conserved_quantities)) ! Total matter (e.g., mol or g)
total = 0
do k=1,nz
do j=1,ny
call model%get_interior_conserved_quantities(1, nx, j, k, total_interior)
do ivar=1,size(model%conserved_quantities)
total(ivar) = total(ivar) + sum(V(:,j,k)*total_interior(:,ivar))
end do
end do
end do
do j=1,ny
call model%get_horizontal_conserved_quantities(1, nx, j, total_interfaces)
do ivar=1,size(model%conserved_quantities)
total(ivar) = total(ivar) + sum(A(:,j)*total_interfaces(:,ivar))
end do
end doAt any time, you can call model%check_interior_state(<LOCATION>, repair, valid), model%check_surface_state(<HORIZONTAL_LOCATION>, repair, valid) and model%check_bottom_state(<HORIZONTAL_LOCATION>, repair, valid) to check the validity of state variable values, and repair these if desired (if repair=.true.). By default, “repairing” implies clipping values to the valid range defined by the biogeochemical model, but a biogeochemical model can also provided custom logic for state variable repair. On return, these routines will have set valid. It will be .true. if the state has been found valid, and .false if not. If valid is .false. and repair is .true., the state will have been repaired (clipped), allowing the simulation to continue.
This routine can both be used to check the validity of the biogeochemical variables, for instance, to adaptively reduce the time step of integration if variable values become invalid, to obtain “corrected” values needed to continue a simulation, or to shut down a simulation where variables take on values outside their prescribed range.
Some common examples:
! 0D box
logical, parameter :: repair = .false.
logical :: valid_int, valid_sf, valid_bt
call model%check_interior_state(repair, valid_int)
call model%check_surface_state(repair, valid_sf)
call model%check_bottom_state(repair, valid_bt)
if (.not. (valid_int .and. valid_sf .and. valid_bt) .and. .not. repair) stop
! 1D water column with extents nz
logical, parameter :: repair = .false.
logical :: valid_int, valid_sf, valid_bt
call model%check_interior_state(1, nz, repair, valid_int)
call model%check_surface_state(repair, valid_sf)
call model%check_bottom_state(repair, valid_bt)
if (.not. (valid_int .and. valid_sf .and. valid_bt) .and. .not. repair) stop
! 3D basin with extents nx,ny,nz
logical, parameter :: repair = .false.
logical :: valid_int, valid_sf, valid_bt
do k=1,nz
do j=1,ny
call model%check_interior_state(1, nx, j, k, repair, valid_int)
if (.not. (valid_int .or. repair)) stop
end do
end do
do j=1,ny
call model%check_surface_state(1, nx, j, repair, valid_sf)
call model%check_bottom_state(1, nx, j, repair, valid_bt)
if (.not. (valid_sf .and. valid_bt) .and. .not. repair) stop
end doTo retrieve the value of the model's interior diagnostic variables, call model%get_interior_diagnostic_data(index). To retrieve the value of the model's horizontal diagnostic variables (e.g., relating to surface or bottom), call model%get_horizontal_diagnostic_data(index). In both cases, index is an integer that specifies the index of the diagnostic variable. This index can range between 1 and size(model%interior_diagnostic_variables) for interior diagnostics, and between 1 and size(model%horizontal_diagnostic_variables) for horizonal diagnostics.
Both routines return a pointer to the diagnostic data, which is contained in an array spanning to entire model domain in the case of get_interior_diagnostic_data, and in an array spanning an horizontal slice of the entire model domain in the case of get_horizontal_diagnostic_data.
Metadata for diagnostic variables are available from arrays model%interior_diagnostic_variables and model%horizontal_diagnostic_variables. Among others, elements of these arrays have members name, long_name, and units (all strings) as well as missing_value (type real(rk)).
Not all diagnostics are computed by default. FABM creates several diagnostics for internal use that are by default not made available to the host. You can tell this from the save attribute of each variable in model%interior_diagnostic_variables and model%horizontal_diagnostic_variables. That attribute is initially .true. for diagnostics defined by biogeochemical models, but .false. for FABM's internal diagnostics. You can however change the value of save (until you call model%start) to tell FABM which particular diagnostics are needed. For instance, you can set save to .true. for internal diagnostics to ensure FABM will compute their value, or you can set save to .false. to tell FABM that it does not need to store or process that diagnostic. For optimal performance, it is recommended to ensure save is .true. only for those diagnostics that you actually need. It should be noted that during a simulation, the value of diagnostics with save=.false. will not be meaningful.
All of the above subroutines either operate on a single location in the spatial domain, or on a 1D slice of the spatial domain. This depends on the host-specific preprocessor definitions that are set. Here the following rules apply:
- if
_FABM_VECTORIZED_DIMENSION_INDEX_is not defined, all subroutines operate on a single point in the domain. The<LOCATION>argument then comprises a set of_FABM_DIMENSION_COUNT_spatial indices that specify the current location (e.g.,kfor a 1D water column,i,j,kfor a 3D model, none for a non-spatial 0D model). Similarly, the<HORIZONTAL_LOCATION>argument taken by horizontal-only routinesget_surface_sources,get_bottom_sourcesandget_horizontal_conserved_quantitiescomprises indices for all non-depth dimensions. That is, it equals<LOCATION>but excludes the index for the depth dimension (if_FABM_DEPTH_DIMENSION_INDEX_is defined). - If
_FABM_VECTORIZED_DIMENSION_INDEX_is defined, all subroutines that related to the pelagic (get_interior_sources,get_vertical_movement,check_interior_state,get_interior_conserved_quantities) operate on a 1D slice of the spatial domain. The<LOCATION>argument then comprises the lower and upper index of range that must be processed for the vectorized dimension, plus constant indices for all other dimensions. For instance,1,nzfor a 1D model, or1,nx,j,kfor a 3D model. If the vectorized dimension is not the same as the depth dimension (or the model does not have a depth dimension), the<HORIZONTAL_LOCATION>argument taken by horizontal-only subroutines (get_surface_sources,get_bottom_sources,get_horizontal_conserved_quantities) has the same structure as<LOCATION>, but it excludes the (constant) index for the depth dimension. If the vectorized dimension is the depth dimension (i.e., if_FABM_VECTORIZED_DIMENSION_INDEX_and_FABM_DEPTH_DIMENSION_INDEX_are both defined and equal), horizontal-only subroutines operate on a single point only, which means that<HORIZONTAL_LOCATION>is comprises the indices for all non-depth dimensions.
Some common cases:
| Model | Preprocessor settings | <LOCATION> |
<HORIZONTAL_LOCATION> |
|---|---|---|---|
| 0D box | _FABM_DIMENSION_COUNT_=0 |
||
| 1D water column |
_FABM_DIMENSION_COUNT_=1_FABM_VECTORIZED_DIMENSION_INDEX_=1_FABM_DEPTH_DIMENSION_INDEX_=1
|
kstart,kstop |
|
| 3D basin |
_FABM_DIMENSION_COUNT_=3_FABM_VECTORIZED_DIMENSION_INDEX_=1_FABM_DEPTH_DIMENSION_INDEX_=3
|
istart,istop,j,k |
istart,istop,j |
Additional information on the model (e.g. long names, units for its variables) is present in members of the model object, after the model has been initialized. This information is located in arrays describing variables per category, e.g., interior_state_variables, bottom_state_variables, surface_state_variables, interior_diagnostic_variables, horizontal_diagnostic_variables, conserved_quantities. Each member of these arrays is a derived type with at minimum the members name, units, long_name, minimum, maximum, missing_value. The host can include this information as metadata in the model output. Further information can be obtained by looking through src/fabm.F90, which defines the derived types (e.g., type_fabm_variable) that hold variable metadata.
integer :: i
do i = 1, size(model%interior_state_variables)
write (*,*) model%interior_state_variables(i)%name, model%interior_state_variables(i)%long_name, model%interior_state_variables(i)%units
end doWhile the host model should not need the parameters of biogeochemical models for any of its computations, it might want to enumerate the parameters to communicate them to the user. For instance, you might want the write the value of the biogeochemical parameters to screen, or save them as part of the model results. This is possible in FABM - you can enumerate all biogeochemical parameters along with their metadata (descriptive name, units):
use fabm_types
use yaml_settings
type (type_model_list_node), pointer :: instance
type (type_key_value_pair), pointer :: pair
! Get pointer to the first model instance
instance => model%root%children%first
do while (associated(instance))
if (instance%model%user_created) then
print *, 'Instance ', trim(instance%model%name)
pair => instance%model%parameters%first
do while (associated(pair))
select type (value => pair%value)
! For each parameter, the following metadata is available:
! - pair%name: short name [string]
! - value%long_name: descriptive name [string]
! - value%units: units [string]
! - value%pvalue: pointer to the parameter value [data type depends on type of "property" object]
! - value%default: default value provided by the BGC model that registered the parameter [data type depends on type of "property" object]
! - value%has_default: whether a default value has been provided during registration [logical]
class is (type_real_setting)
print *, ' ', pair%name, ': ', value%pvalue
class is (type_integer_setting)
print *, ' ', pair%name, ': ', value%pvalue
class is (type_logical_setting)
print *, ' ', pair%name, ': ', value%pvalue
class is (type_string_setting)
print *, ' ', pair%name, ': ', value%pvalue
end select
pair => pair%next
end do
end if
instance => instance%next
end doFor questions about FABM's use or development, visit Discussions. If you would like to cite FABM, please refer to its main publication and/or URLs.
Background
User guide
- Obtaining the source code
- Building and installing
- Setting up a simulation
- Available biogeochemical models
- Specific hosts
Developer guide
Updates
Tips and tricks
Support
How to cite
Licensing and copyright
Acknowledgements
Presentations
Workshops