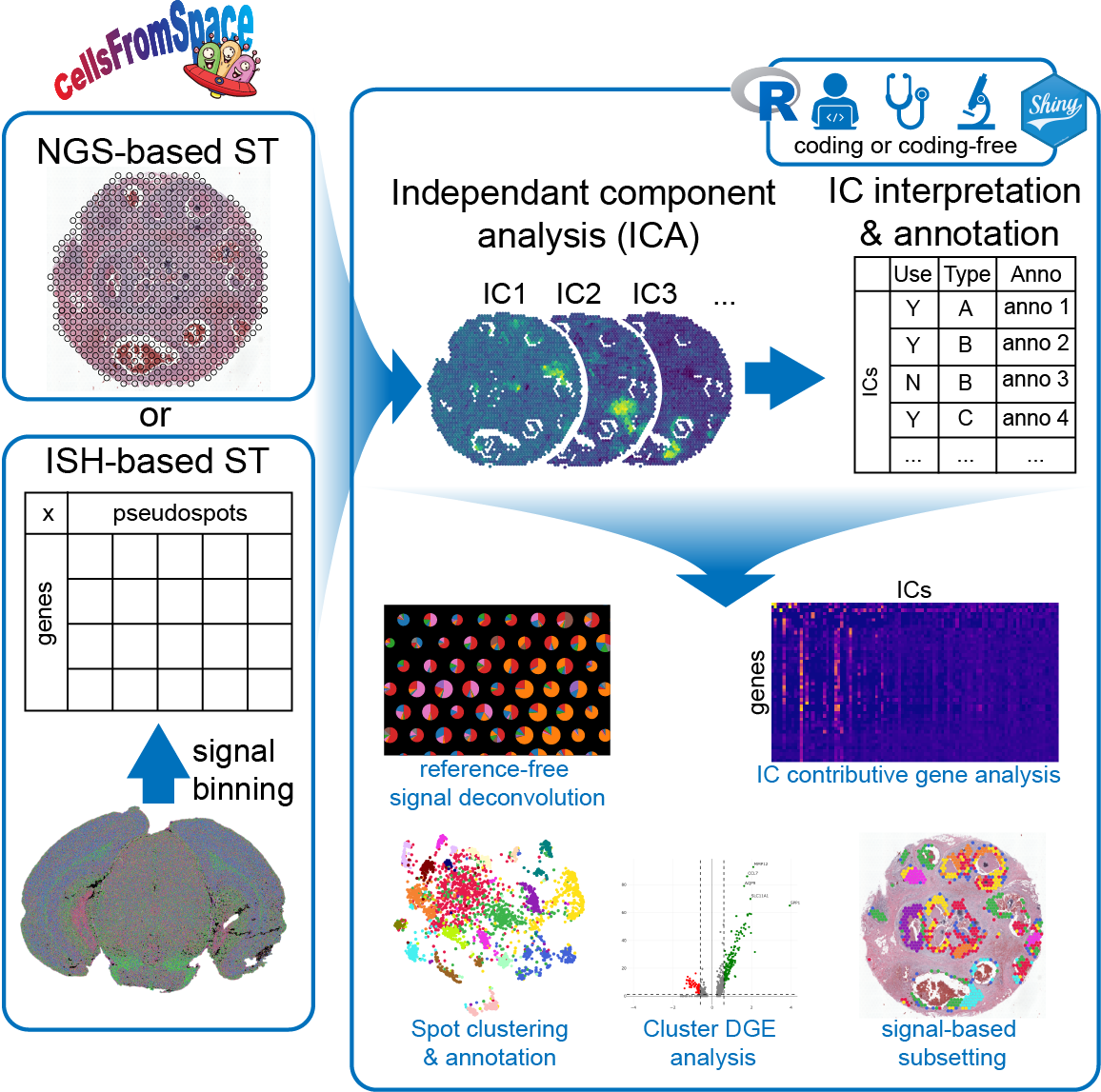
Built on the Seurat ecosystem, CellsFromSpace (CFS) is an R package designed for user-friendly spatial transcriptomics (ST) analysis using independent component analysis (ICA), enabling reference-free signal decomposition and spot deconvolution through interpretable latent space.
Full documentation and tutorial can be found at: Tutorial and instructions
More documentation and tutorials are being made as new features are implemented. The tool can already be installed and is still un beta but is being actively worked on for full release.
CellsFromSpace aims at providing a simple and user-friendly ST analysis pipeline to any researcher, biologist, pathologist or bioinformatician regardless of coding ability. As a reference-free complete analysis tool, any dataset can be processed and analysed from end to end within the CFS interactive interface. CFS also allows integrated analysis of multiple samples for complex datasets.
CellsFromSpace can be installed from the github repository using the package devtools :
install.packages("devtools")
devtools::install_github("gustaveroussy/CFS"
,ref="main")To install the development branch with additional (potentially unstable) features, use:
devtools::install_github("gustaveroussy/CFS"
,ref="development")CellsFromSpace contains functions for the processing of Spatial Transcriptomics (ST) data from major commercial technologies built into a fully interactive Shiny interface. Most pre-processing functionalities and all processing, analysis and visualization functionalities of CFS can be accessed through the interactive shiny interface, providing an all-in-one easy-to-use analytical tool for ST data.
Once installed, simply launch the interactive Shiny interface to access all of CFS' functionalities:
library(CFS)
library(Seurat)
launchCFS()Follow along our [Tutorials](Tutorial URL) for a step-by-step exploration of the CFS pipeline. Briefly, the analysis workflow consists of:
- Pre-processing
- IC annotation*
- Visualization and Analysis
*IC annotation is the crux of the CFS pipeline. Multiple tools are implemented to assist scientists in this crucial interpretation step
This work is published in Bioinformatics Advances. You can cite the article as:
@article{10.1093/bioadv/vbae081,
author = {Thuilliez, Corentin and Moquin-Beaudry, Gaël et al.},
title = "{CellsFromSpace: a fast, accurate, and reference-free tool to deconvolve and annotate spatially distributed omics data}",
journal = {Bioinformatics Advances},
volume = {4},
number = {1},
pages = {vbae081},
year = {2024},
month = {05},
issn = {2635-0041},
doi = {10.1093/bioadv/vbae081},
url = {https://doi.org/10.1093/bioadv/vbae081},
eprint = {https://academic.oup.com/bioinformaticsadvances/article-pdf/4/1/vbae081/58310899/vbae081.pdf},
}
This work is distributed under the the terms of the creative commons attribution Licence Creative Commons Attribution - No commercial use - shared in the same conditions 4.0 International.