-
Notifications
You must be signed in to change notification settings - Fork 0
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
1 changed file
with
234 additions
and
2 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,4 +1,236 @@ | ||
| # cotengrust | ||
|
|
||
| `cotengrust` provides fast rust implemented versions of contraction ordering | ||
| primitives for tensor networks and einsums. | ||
| `cotengrust` provides fast rust implementations of contraction ordering | ||
| primitives for tensor networks or einsum expressions. The two main functions | ||
| are: | ||
|
|
||
| - `optimize_optimal(inputs, output, size_dict, **kwargs)` | ||
| - `optimize_greedy(inputs, output, size_dict, **kwargs)` | ||
|
|
||
| The optimal algorithm is an optimized version of the `opt_einsum` 'dp' | ||
| path - itself an implementation of https://arxiv.org/abs/1304.6112. | ||
|
|
||
|
|
||
| ## Installation | ||
|
|
||
| `cotengrust` is available for most platforms from | ||
| [PyPI](https://pypi.org/project/cotengrust/): | ||
|
|
||
| ```bash | ||
| pip install cotengrust | ||
| ``` | ||
|
|
||
| or if you want to develop locally (which requires [pyo3](https://github.com/PyO3/pyo3) | ||
| and [maturin](https://github.com/PyO3/maturin)): | ||
|
|
||
| ```bash | ||
| git clone https://github.com/jcmgray/cotengrust.git | ||
| cd cotengrust | ||
| maturin develop --release | ||
| ``` | ||
| (the release flag is very important for assessing performance!). | ||
|
|
||
|
|
||
| ## Usage | ||
|
|
||
| If `cotengrust` is installed, then by default `cotengra` will use it for its | ||
| greedy and optimal subroutines, notably subtree reconfiguration. You can also | ||
| call the routines directly: | ||
|
|
||
| ```python | ||
| import cotengra as ctg | ||
| import cotengrust as ctgr | ||
|
|
||
| # specify an 8x8 square lattice contraction | ||
| inputs, output, shapes, size_dict = ctg.utils.lattice_equation([8, 8]) | ||
|
|
||
| # find the optimal 'combo' contraction path | ||
| %%time | ||
| path = ctgr.optimize_optimal(inputs, output, size_dict, minimize='combo') | ||
| # CPU times: user 13.7 s, sys: 83.4 ms, total: 13.7 s | ||
| # Wall time: 13.7 s | ||
|
|
||
| # construct a contraction tree for further introspection | ||
| tree = ctg.ContractionTree.from_path( | ||
| inputs, output, size_dict, path=path | ||
| ) | ||
| tree.plot_rubberband() | ||
| ``` | ||
| 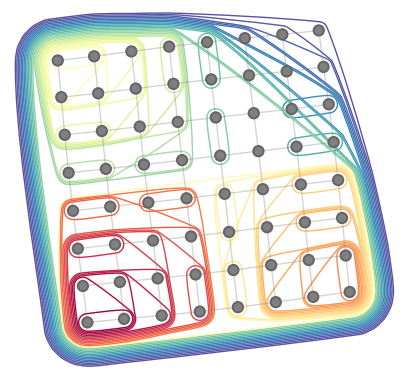 | ||
|
|
||
|
|
||
| ## API | ||
|
|
||
| The optimize functions follow the api of the python implementations in `cotengra.pathfinders.path_basic.py`. | ||
|
|
||
| ```python | ||
| def optimize_optimal( | ||
| inputs, | ||
| output, | ||
| size_dict, | ||
| minimize='flops', | ||
| cost_cap=2, | ||
| search_outer=False, | ||
| simplify=True, | ||
| use_ssa=False, | ||
| ): | ||
| """Find an optimal contraction ordering. | ||
| Parameters | ||
| ---------- | ||
| inputs : Sequence[Sequence[str]] | ||
| The indices of each input tensor. | ||
| output : Sequence[str] | ||
| The indices of the output tensor. | ||
| size_dict : dict[str, int] | ||
| The size of each index. | ||
| minimize : str, optional | ||
| The cost function to minimize. The options are: | ||
| - "flops": minimize with respect to total operation count only | ||
| (also known as contraction cost) | ||
| - "size": minimize with respect to maximum intermediate size only | ||
| (also known as contraction width) | ||
| - 'write' : minimize the sum of all tensor sizes, i.e. memory written | ||
| - 'combo' or 'combo={factor}` : minimize the sum of | ||
| FLOPS + factor * WRITE, with a default factor of 64. | ||
| - 'limit' or 'limit={factor}` : minimize the sum of | ||
| MAX(FLOPS, alpha * WRITE) for each individual contraction, with a | ||
| default factor of 64. | ||
| 'combo' is generally a good default in term of practical hardware | ||
| performance, where both memory bandwidth and compute are limited. | ||
| cost_cap : float, optional | ||
| The maximum cost of a contraction to initially consider. This acts like | ||
| a sieve and is doubled at each iteration until the optimal path can | ||
| be found, but supplying an accurate guess can speed up the algorithm. | ||
| search_outer : bool, optional | ||
| If True, consider outer product contractions. This is much slower but | ||
| theoretically might be required to find the true optimal 'flops' | ||
| ordering. In practical settings (i.e. with minimize='combo'), outer | ||
| products should not be required. | ||
| simplify : bool, optional | ||
| Whether to perform simplifications before optimizing. These are: | ||
| - ignore any indices that appear in all terms | ||
| - combine any repeated indices within a single term | ||
| - reduce any non-output indices that only appear on a single term | ||
| - combine any scalar terms | ||
| - combine any tensors with matching indices (hadamard products) | ||
| Such simpifications may be required in the general case for the proper | ||
| functioning of the core optimization, but may be skipped if the input | ||
| indices are already in a simplified form. | ||
| use_ssa : bool, optional | ||
| Whether to return the contraction path in 'single static assignment' | ||
| (SSA) format (i.e. as if each intermediate is appended to the list of | ||
| inputs, without removals). This can be quicker and easier to work with | ||
| than the 'linear recycled' format that `numpy` and `opt_einsum` use. | ||
| Returns | ||
| ------- | ||
| path : list[list[int]] | ||
| The contraction path, given as a sequence of pairs of node indices. It | ||
| may also have single term contractions if `simplify=True`. | ||
| """ | ||
| ... | ||
|
|
||
|
|
||
| def optimize_greedy( | ||
| inputs, | ||
| output, | ||
| size_dict, | ||
| costmod=1.0, | ||
| temperature=0.0, | ||
| simplify=True, | ||
| use_ssa=False, | ||
| ): | ||
| """Find a contraction path using a (randomizable) greedy algorithm. | ||
| Parameters | ||
| ---------- | ||
| inputs : Sequence[Sequence[str]] | ||
| The indices of each input tensor. | ||
| output : Sequence[str] | ||
| The indices of the output tensor. | ||
| size_dict : dict[str, int] | ||
| A dictionary mapping indices to their dimension. | ||
| costmod : float, optional | ||
| When assessing local greedy scores how much to weight the size of the | ||
| tensors removed compared to the size of the tensor added:: | ||
| score = size_ab - costmod * (size_a + size_b) | ||
| This can be a useful hyper-parameter to tune. | ||
| temperature : float, optional | ||
| When asessing local greedy scores, how much to randomly perturb the | ||
| score. This is implemented as:: | ||
| score -> sign(score) * log(|score|) - temperature * gumbel() | ||
| which implements boltzmann sampling. | ||
| simplify : bool, optional | ||
| Whether to perform simplifications before optimizing. These are: | ||
| - ignore any indices that appear in all terms | ||
| - combine any repeated indices within a single term | ||
| - reduce any non-output indices that only appear on a single term | ||
| - combine any scalar terms | ||
| - combine any tensors with matching indices (hadamard products) | ||
| Such simpifications may be required in the general case for the proper | ||
| functioning of the core optimization, but may be skipped if the input | ||
| indices are already in a simplified form. | ||
| use_ssa : bool, optional | ||
| Whether to return the contraction path in 'single static assignment' | ||
| (SSA) format (i.e. as if each intermediate is appended to the list of | ||
| inputs, without removals). This can be quicker and easier to work with | ||
| than the 'linear recycled' format that `numpy` and `opt_einsum` use. | ||
| Returns | ||
| ------- | ||
| path : list[list[int]] | ||
| The contraction path, given as a sequence of pairs of node indices. It | ||
| may also have single term contractions if `simplify=True`. | ||
| """ | ||
|
|
||
| def optimize_simplify( | ||
| inputs, | ||
| output, | ||
| size_dict, | ||
| use_ssa=False, | ||
| ): | ||
| """Find the (partial) contracton path for simplifiactions only. | ||
| Parameters | ||
| ---------- | ||
| inputs : Sequence[Sequence[str]] | ||
| The indices of each input tensor. | ||
| output : Sequence[str] | ||
| The indices of the output tensor. | ||
| size_dict : dict[str, int] | ||
| A dictionary mapping indices to their dimension. | ||
| use_ssa : bool, optional | ||
| Whether to return the contraction path in 'single static assignment' | ||
| (SSA) format (i.e. as if each intermediate is appended to the list of | ||
| inputs, without removals). This can be quicker and easier to work with | ||
| than the 'linear recycled' format that `numpy` and `opt_einsum` use. | ||
| Returns | ||
| ------- | ||
| path : list[list[int]] | ||
| The contraction path, given as a sequence of pairs of node indices. It | ||
| may also have single term contractions. | ||
| """ | ||
| ... | ||
|
|
||
| def ssa_to_linear(ssa_path, n=None): | ||
| """Convert a SSA path to linear format.""" | ||
| ... | ||
|
|
||
| def find_subgraphs(inputs, output, size_dict,): | ||
| """Find all disconnected subgraphs of a specified contraction.""" | ||
| ... | ||
| ``` | ||
|
|