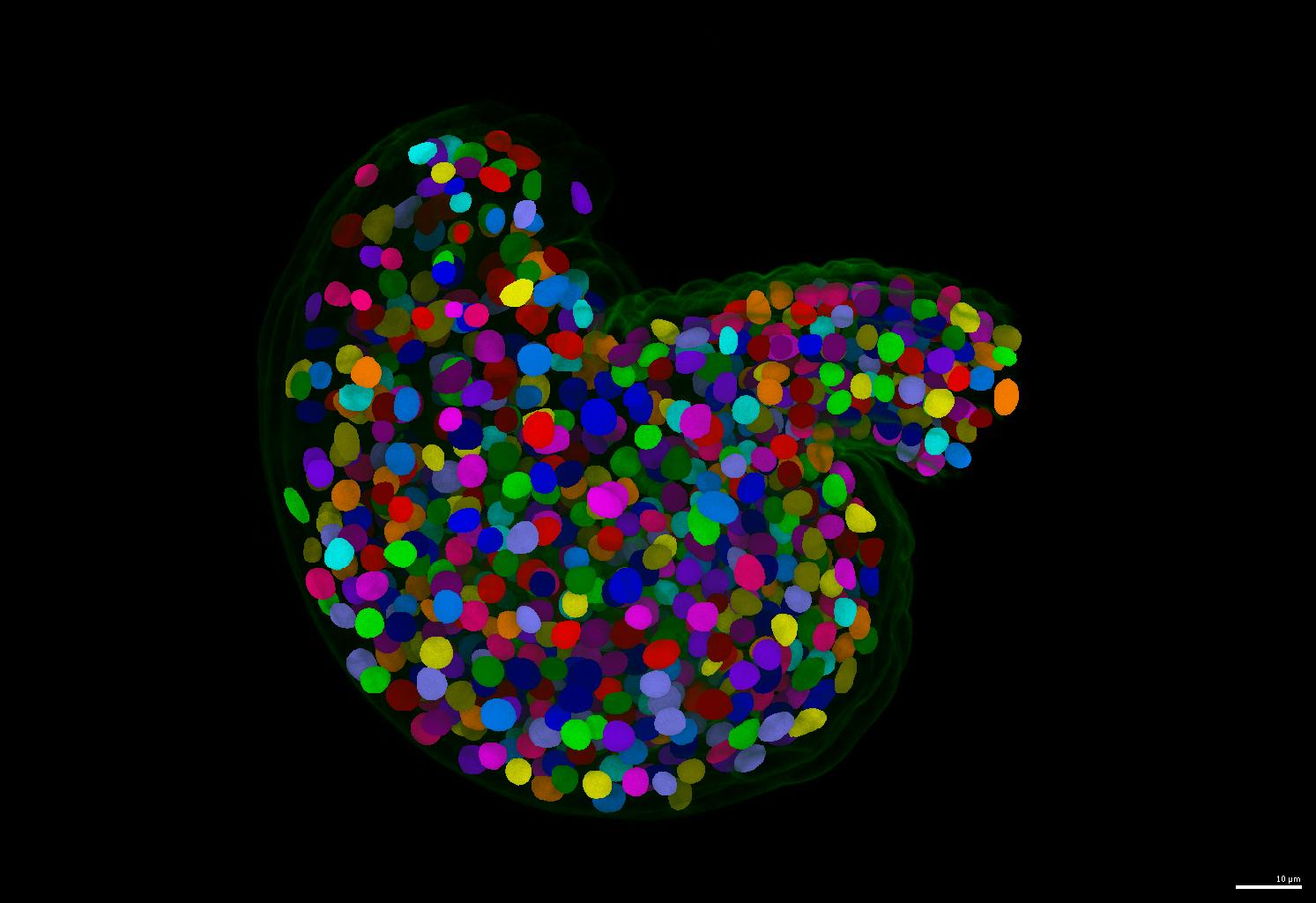
The GoNuclear repository hosts the code and guides for the pipelines used in the paper A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context. It is structured in to four folders:
- stardist/ contains a 3D StarDist training and inference pipeline,
run-stardist. - plantseg/ contains configuration files for training and inference with PlantSeg.
- cellpose/ contains scripts for training and inference with Cellpose.
- evaluation/ contains modules for evaluating the segmentation results.
and are described in GoNuclear documentation 📖.
Please go to BioImage Archive S-BIAD1026 for the training data and models. I organised them in the following structure:
Training data
├── 2d/
│ ├── isotropic/
│ │ ├── gold/
│ │ └── initial/
│ └── original/
│ ├── gold/
│ └── README.txt
└── 3d_all_in_one/
├── 1135.h5
├── 1136.h5
├── 1137.h5
├── 1139.h5
└── 1170.h5
Models
├── cellpose/
│ ├── cyto2_finetune/
│ │ └── gold/
│ ├── nuclei_finetune/
│ │ ├── gold/
│ │ └── initial/
│ └── scratch_trained/
│ └── gold/
├── plantseg/
│ └── 3dunet/
│ ├── gold/
│ ├── initial/
│ ├── platinum/
│ └── train_example.yml
└── stardist/
├── resnet/
│ ├── gold/
│ ├── initial/
│ └── platinum/
├── train_example.yml
└── unet/
└── gold/If you find this work useful, please cite our paper and the respective tools' papers:
@article{vijayan2024deep,
title={A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context},
author={Vijayan, Athul and Mody, Tejasvinee Atul and Yu, Qin and Wolny, Adrian and Cerrone, Lorenzo and Strauss, Soeren and Tsiantis, Miltos and Smith, Richard S and Hamprecht, Fred A and Kreshuk, Anna and others},
journal={Development},
volume={151},
number={14},
year={2024},
publisher={The Company of Biologists}
}