-
Notifications
You must be signed in to change notification settings - Fork 4
Home
DiscoPlot allows the user to quickly identify genomic rearrangements, misassemblies and sequencing artefacts by providing a scalable method for visualising large sections of the genome. It reads paired-reads alignments in SAM or BAM format and single-end reads standard BLAST tab format and creates a scatter plot of opaque crosses representing the alignments to a reference. DiscoPlot is freely available (under a GPL license) for download (Mac OS X, Unix and Windows).
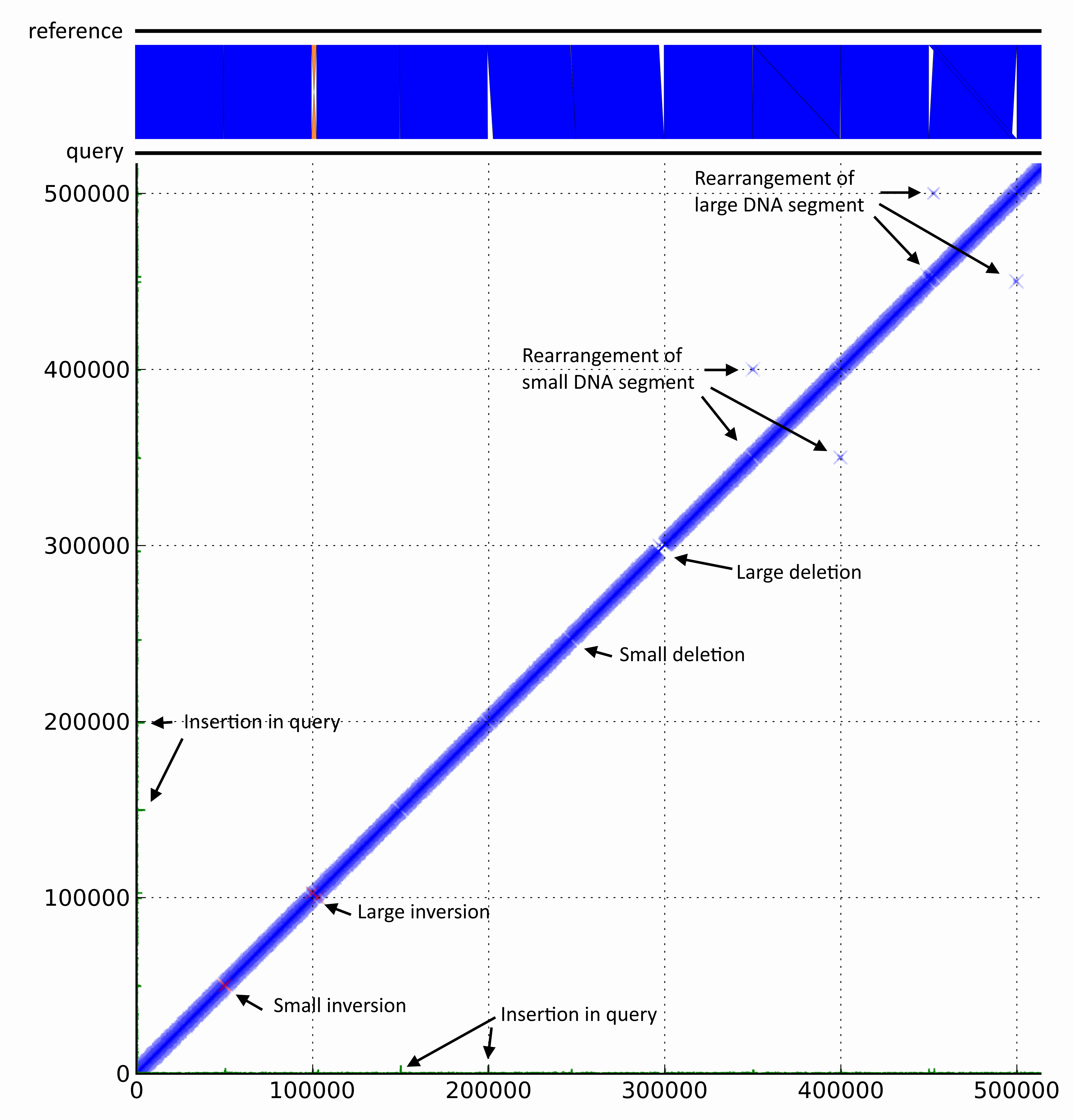
DiscoPlot of a mock genome. A mock genome was created by adding genomic rearrangements to the chromosome of E. coli str. UTI89. Paired-end reads generated from the mock genome (query) with GemSim and mapped back to UTI89 (reference). The first ~500 Kbp were then visualised using DiscoPlot.