Adaptive is an open-source Python library that streamlines adaptive parallel function evaluations. Rather than calculating all points on a dense grid, it intelligently selects the "best" points in the parameter space based on your provided function and bounds. With minimal code, you can perform evaluations on a computing cluster, display live plots, and optimize the adaptive sampling algorithm.
Adaptive is most efficient for computations where each function evaluation takes at least ≈50ms due to the overhead of selecting potentially interesting points.
To see Adaptive in action, try the example notebook on Binder or explore the tutorial on Read the Docs.
[ToC] 📚
- 🎯 Intelligent Adaptive Sampling: Adaptive focuses on areas of interest within a function, ensuring better results with fewer evaluations, saving time, and computational resources.
- ⚡ Parallel Execution: The library leverages parallel processing for faster function evaluations, making optimal use of available computational resources.
- 📊 Live Plotting and Info Widgets: When working in Jupyter notebooks, Adaptive offers real-time visualization of the learning process, making it easier to monitor progress and identify areas of improvement.
- 🔧 Customizable Loss Functions: Adaptive supports various loss functions and allows customization, enabling users to tailor the learning process according to their specific needs.
- 📈 Support for Multidimensional Functions: The library can handle functions with scalar or vector outputs in one or multiple dimensions, providing flexibility for a wide range of problems.
- 🧩 Seamless Integration: Adaptive offers a simple and intuitive interface, making it easy to integrate with existing Python projects and workflows.
- 💾 Flexible Data Export: The library provides options to export learned data as NumPy arrays or Pandas DataFrames, ensuring compatibility with various data processing tools.
- 🌐 Open-Source and Community-Driven: Adaptive is an open-source project, encouraging contributions from the community to continuously improve and expand the library's features and capabilities.
Adaptively learning a 1D function and live-plotting the process in a Jupyter notebook:
from adaptive import notebook_extension, Runner, Learner1D
notebook_extension()
def peak(x, a=0.01):
return x + a**2 / (a**2 + x**2)
learner = Learner1D(peak, bounds=(-1, 1))
runner = Runner(learner, loss_goal=0.01)
runner.live_info()
runner.live_plot()You can export the learned data as a NumPy array:
data = learner.to_numpy()If you have Pandas installed, you can also export the data as a DataFrame:
df = learner.to_dataframe()The core concept in adaptive is the learner.
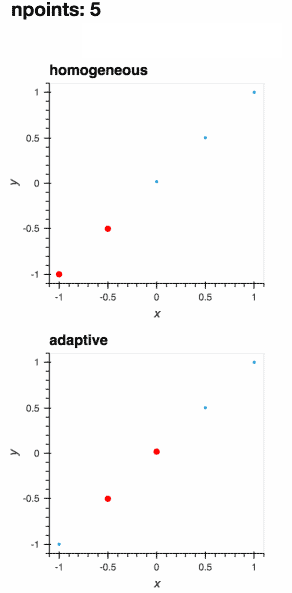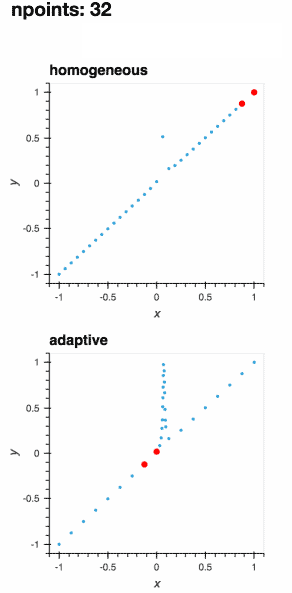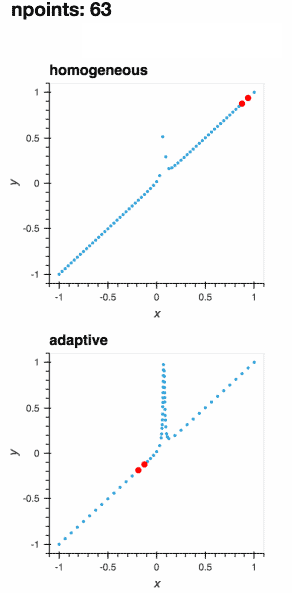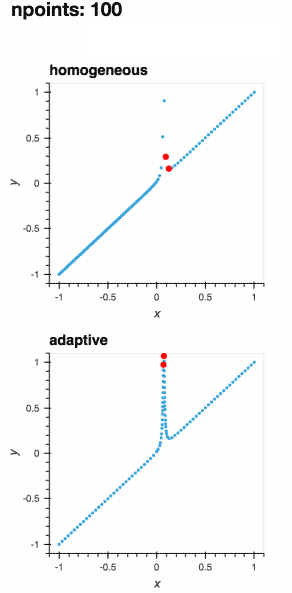
A learner samples a function at the most interesting locations within its parameter space, allowing for optimal sampling of the function.
As the function is evaluated at more points, the learner improves its understanding of the best locations to sample next.
The definition of the "best locations" depends on your application domain.
While adaptive provides sensible default choices, the adaptive sampling process can be fully customized.
The following learners are implemented:
Learner1D: for 1D functionsf: ℝ → ℝ^N,Learner2D: for 2D functionsf: ℝ^2 → ℝ^N,LearnerND: for ND functionsf: ℝ^N → ℝ^M,AverageLearner: for random variables, allowing averaging of results over multiple evaluations,AverageLearner1D: for stochastic 1D functions, estimating the mean value at each point,IntegratorLearner: for integrating a 1D functionf: ℝ → ℝ,BalancingLearner: for running multiple learners simultaneously and selecting the "best" one as more points are gathered.
Meta-learners (to be used with other learners):
BalancingLearner: for running several learners at once, selecting the "most optimal" one each time you get more points,DataSaver: for when your function doesn't return just a scalar or a vector.
In addition to learners, adaptive offers primitives for parallel sampling across multiple cores or machines, with built-in support for:
concurrent.futures,
mpi4py,
loky,
ipyparallel, and
distributed.
adaptive works with Python 3.7 and higher on Linux, Windows, or Mac, and provides optional extensions for working with the Jupyter/IPython Notebook.
The recommended way to install adaptive is using conda:
conda install -c conda-forge adaptiveadaptive is also available on PyPI:
pip install "adaptive[notebook]"The [notebook] above will also install the optional dependencies for running adaptive inside a Jupyter notebook.
To use Adaptive in Jupyterlab, you need to install the following labextensions.
jupyter labextension install @jupyter-widgets/jupyterlab-manager
jupyter labextension install @pyviz/jupyterlab_pyvizClone the repository and run pip install -e ".[notebook,testing,other]" to add a link to the cloned repo into your Python path:
git clone git@github.com:python-adaptive/adaptive.git
cd adaptive
pip install -e ".[notebook,testing,other]"We recommend using a Conda environment or a virtualenv for package management during Adaptive development.
To avoid polluting the history with notebook output, set up the git filter by running:
python ipynb_filter.pyin the repository.
To maintain consistent code style, we use pre-commit. Install it by running:
pre-commit installin the repository.
If you used Adaptive in a scientific work, please cite it as follows.
@misc{Nijholt2019,
doi = {10.5281/zenodo.1182437},
author = {Bas Nijholt and Joseph Weston and Jorn Hoofwijk and Anton Akhmerov},
title = {\textit{Adaptive}: parallel active learning of mathematical functions},
publisher = {Zenodo},
year = {2019}
}If you're interested in the scientific background and principles behind Adaptive, we recommend taking a look at the draft paper that is currently being written. This paper provides a comprehensive overview of the concepts, algorithms, and applications of the Adaptive library.
We would like to give credits to the following people:
- Pedro Gonnet for his implementation of CQUAD, “Algorithm 4” as described in “Increasing the Reliability of Adaptive Quadrature Using Explicit Interpolants”, P. Gonnet, ACM Transactions on Mathematical Software, 37 (3), art. no. 26, 2010.
- Pauli Virtanen for his
AdaptiveTriSamplingscript (no longer available online since SciPy Central went down) which served as inspiration for theadaptive.Learner2D.
For general discussion, we have a Gitter chat channel. If you find any bugs or have any feature suggestions please file a GitHub issue or submit a pull request.