If you are interested, let is know by filling out this form https://forms.gle/UNFiZG9ibDpz82nj8.
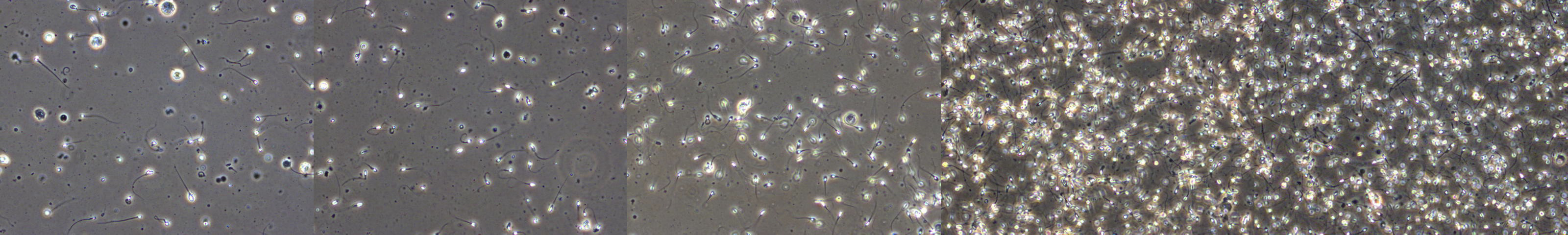
The 2020 BioMedia Grand Challenge tackles the challenge of predicting certain quality measurements of human sperm using a multimodal dataset consisting of microscopic video recordings of human semen, associated sensor data, and participant-related data. Assessment of semen is commonly performed during the early stages of investigating male infertility. This is mostly a manual process, where trained clinicians inspect semen samples through a microscope and count the number of different sperms (such as the number of moving or not moving sperm) to evaluate the quality. As one might expect, this is a tedious and time-consuming process that could greatly benefit from some automation. Furthermore, due to the subjective nature of manually inspecting large numbers of moving sperm, there is large inter- intra-observer variability between and within clinics. Having an algorithm that can give consistent results for any given semen sample would be of great benefit. This year, we present four different tasks, each targeting a different aspect of sperm quality assessment. The first two tasks relate to predicting common measurements used for semen quality assessment, specifically the motility (movement) and morphology (shape and structure) of the spermatozoa (living sperm). These two tasks encourage participants to combine all the available data modalities and sources to make predictions. The third task relates to looking at individual sperm to figure out which one moves the fastest. For all tasks, we require participants to measure and report the data processing performance in terms of time spent on each frame being analyzed. As we aim for real-time applications, the processing speed is an essential factor, especially for the individual spermatozoon selection task during the in vitro fertilization procedure (a procedure where a sperm is injected into an egg outside of the body).
Medical domain experts are included in the organization and evaluation of the results. Medical and industry experts will also be present at the ACM MM conference to provide direct feedback and possibilities to interact with researchers directly. It is a typical assumption that visual analysis, as it is already provided by the computer vision and medical image processing communities today, is capable of already providing viable and practical approaches to healthcare multimedia challenges. Although we concede that these methods are indeed essential contributors to promising approaches, we realize the limitations of analyzing images and videos alone in medical fields such as endoscopy, ultrasound, etc., because of the complexity and needs of medical experts and patients. Neither does it make enough use of the multitude of additional information sources, including sensors and temporal information. The challenge can be seen as very challenging and hard to solve and evaluate. Due to its novel use case, we hope to motivate a lot of researchers to have a look into the field of medical multimedia. Performing research that can have a societal impact will be an essential part of multimedia research in the future. We hope that the challenge can help to raise awareness of the topic and also provide exciting and meaningful use cases to researchers.
The development dataset for the task is the VISEM dataset containing data from 85 male participants aged 18 years or older. For each participant, we include a set of measurements from a standard semen analysis, a video of live spermatozoa, a sperm fatty acid profile, the fatty acid composition of serum phospholipids, study participants-related data and WHO analysis data. The dataset contains over 35 gigabytes of videos, with each video lasting between two to seven minutes. Each video has a resolution of 640 x 480 and runs at 50 frames-per-second. The dataset contains in total six CSV-files (five for data and one which maps video IDs to study participants IDs), a description file, and a folder containing the videos. The name of each video file contains the videos ID, the date it was recorded, and a small optional description. Then, the end of the filename contains the code of the person who assessed the video. Furthermore, VISEM contains five CSV-files for each of the other data provided, a CSV-file with the IDs linked to each their video, and a text file containing descriptions of some of the columns of the CSV-files. One row in each CSV-file represents a participant. The provided CSV-files are:
- semen_analysis_data.csv: The results of standard semen analysis.
- fatty_acids_spermatozoa.csv: The levels of several fatty acids in the spermatozoa of the participants.
- fatty_acids_serum.csv: The serum levels of the fatty acids of the phospholipids (measured from the blood of the participant).
- sex_hormones.csv: The serum levels of sex hormones measured in the blood of the participants.
- study_participant_related_data.csv: General information about the participants such as age, abstinence time and Body Mass Index (BMI).
- videos.csv: Overview of which video-file belongs to what participants.
We will provide pre-extracted features for all videos in the dataset. For the final evaluation, participants will use a provided test dataset without annotations. The VISEM dataset is publicly available for participants and other multimedia researchers without any restriction. All study participants agreed to donate their data for science and provided the necessary consent for us to be able to distribute the data (checked and approved by Norwegian data authority and ethical committee). You can find the dataset at the following link datasets.simula.no/visem.
To participate, each participants will have to submit runs for the following two tasks:
- Prediction of motility. The goal of this task is to predict the percentage of progressive, non-progressive, and immotile sperm in a given video sample. Motility is the ability of an organism to move independently. Where a progressive spermatozoon can "move forward", a non-progressive would move in circles without progression. The prediction needs to be performed sample wise resulting in one set of predictions per video sample. No sperm tracking or bounding boxes are required to solve the task.
- Prediction of morphology. Morphology is a branch of biology dealing with the study of an organism's form and structural features. In the context of semen, doctors often examine the three parts which make up a spermatozoon, which includes the head, midpiece, and tail. This task should predict the percentage of sperm with head defects, midpiece defects, and tail defects. Morphology analysis also only requires sample wise prediction resulting in one value per sample per predicted attribute. Similar to the motility task, no sperm tracking or bounding boxes are required to solve the task.
For both tasks, participants are asked to perform video analysis over single frame analysis. This is important since single frame-based analysis will not be able to catch the movement of the spermatozoa, which contains essential information to perform the predictions.
The two optional sub-tasks are:
- Unsupervised sperm tracking. Find the spermatozoon that moves fastest compared to all others. This task requires that task participants track the spermatozoa. Within the tracked ones, the fastest is defined as:
- Fastest average speed - the one that moves the longest distance during the video (total distance/length of the video). This can then be calculated summarizing the different positions (in pixels) between each frame and divide on the number of frames.
- Highest top speed - the one that has the highest average speed. This can be calculated using the maximum of the differences between frames. One specific challenge with this subtask is that the video also changes the view on the sample. This happens because the sample is moved below the microscope to observe the complete sample area. Therefore, the tracking has to be performed per viewpoint on the sample.
- Sperm tracking support tool. Participants are asked to come up with a tool that can assist medical experts better in observing and choosing sperms. This could be as simple as providing an interface with different filters that allow for better control of the tracking of spermatozoa. The quality of the tool is decided via a majority vote from three medical experts in the field testing the tool. The assessment will focus on the usefulness and novelty of the idea (not user interface or usability in terms of human-computer interaction).
For the evaluation, we will use mean squared error, mean absolute error and the mean absolute percentage error for first two challenges. For the ranking mean absolute percentage error will be used which shows the improvement of the automatic prediction compared to the average prediction. For the optional challenges, we will use manual evaluation with the help from three different experts within human reproduction.
- Steven Hicks, steven@simula.no, SimulaMet & OsloMet
- Pål Halvorsen, paalh@simula.no, SimulaMet & OsloMet
- Michael Riegler, michael@simula.no, SimulaMet
- Vajira Thambawita, vajira@simula.no, SimulaMet & OsloMet
- Hugo L. Hammer, hugo@simula.no, SimulaMet & OsloMet
- Trine B. Haugen, tribha@oslomet.no, Oslo Metropolitan University
If you need help or have any questions please contact Steven Hicks, Pål Halvorsen, or Michael Riegler. Please mark the email with ACM Grand Challenge Biomedia 2020.