This project is a tool to build CheXNet-like models, written in Keras.
What is CheXNet?
ChexNet is a deep learning algorithm that can detect and localize 14 kinds of diseases from chest X-ray images. As described in the paper, a 121-layer densely connected convolutional neural network is trained on ChestX-ray14 dataset, which contains 112,120 frontal view X-ray images from 30,805 unique patients. The result is so good that it surpasses the performance of practicing radiologists.
- Train/test a baseline model by following the quickstart. You can get a model with performance close to the paper.
- Modify
multiplyanduse_class_balancingparameters inconfig.inito see if you can get better performance. - Modify
weights.pyto customize your weights in loss function. - Every time you do a new experiment, make sure you modify
output_dirinconfig.iniotherwise previous training results might be overwritten. For more options check the parameter description inconfig.ini.
Note that currently this project can only be executed in Linux and macOS. You might run into some issues in Windows.
- Download all tar files and Data_Entry_2017.csv of ChestX-ray14 dataset from NIH dropbox. Put them under
./datafolder and untar all tar files. - Download DenseNet-121 ImageNet tensorflow pretrained weights from DenseNet-Keras. Specify the file path in
config.ini(field:base_model_weights_file) - Create & source a new virtualenv. Python >= 3.6 is required.
- Install dependencies by running
pip3 install -r requirements.txt. - Run
./kmi_train <config_file>.inito train a new model. If you want to run the training using multiple GPUs, just prependCUDA_VISIBLE_DEVICES=0,1,...to restrict the GPU devices.nvidia-smicommand will be helpful if you don't know which device are available. - Run
./kmi_test <config_file>.inito test your trained model
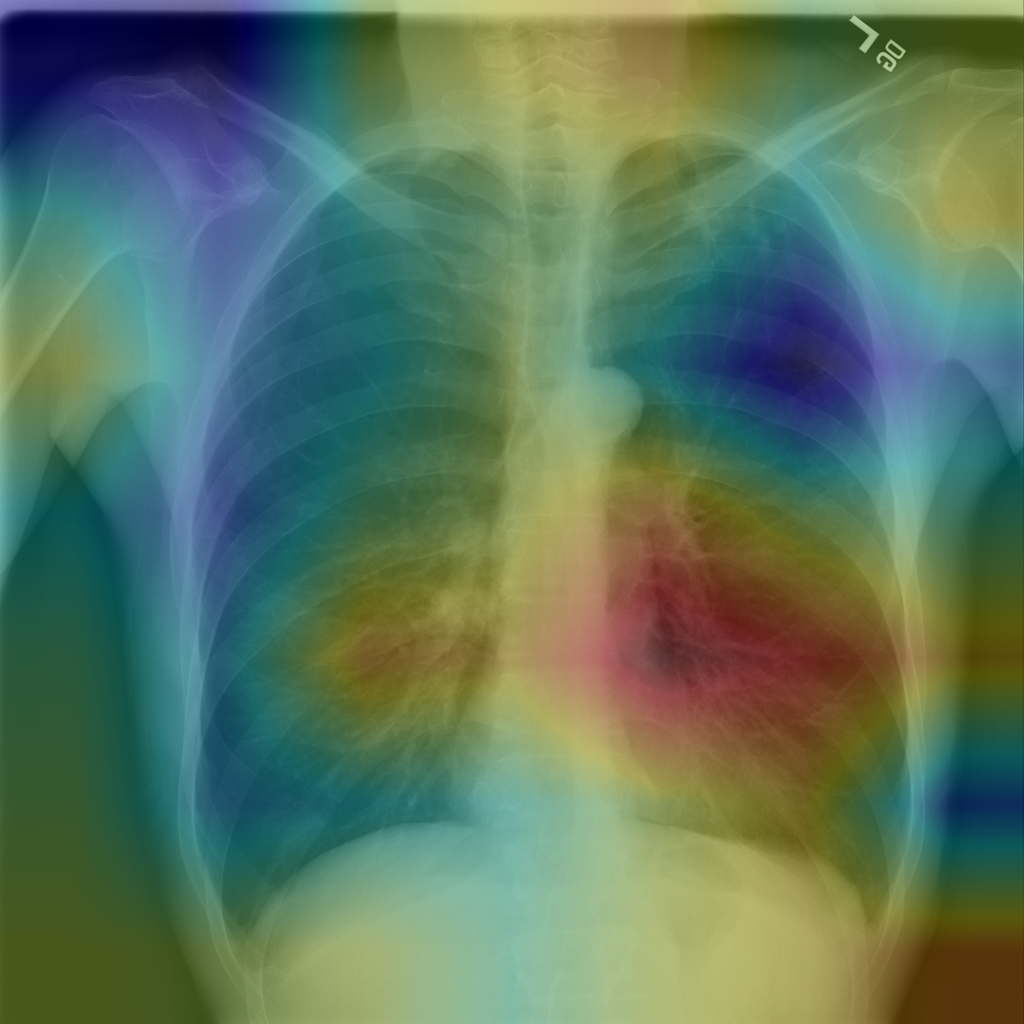
Reference: Grad-CAM. CAM image is generated as accumumlated weighted activation before last global average pooling (GAP) layer. It is scaled up to dimensions of original image.
To enable this feature during testing, edit config.ini as follows:
; ... (lines omitted) ...
[TEST]
; ... (lines omitted) ...
; Save grad-cam outputs
enable_grad_cam = true
; ... (lines omitted) ...
Make sure enable_grad_cam = true is properly set under section [TEST]
Execute following command in your shell to initiate model testing
./kmi_test <config_file_name>.ini
CAM images will be generated into $pwd/imgdir.
Note: Guided back-prop is still a TODO item.
Two configuration files are provided as default setup
- chexnet_config.ini : Configureation for CXR8 dataset (
used in CheXNet) - pxr_config.ini (The hip fracture dataset)
[DEFAULT]
; working directory, one working directory can only have one running job at a time
output_dir = ./experiments/pxr0
; Verbosity for other stuffs
verbosity = 0
; all images should be placed under this dir
image_source_dir = ./datasets/pxr_ds/images/
; Fully Connected model type [multiclass, multibinary]
class_mode = multiclass
; number of train/dev unique patient count,
; test patient count will be (total count - train_patient_count - dev_patient_count), which is 389 by default
train_patient_ratio = 75
dev_patient_ratio = 5
; 512 means 512x512 pixels, always use square shaped input
image_dimension = 256
; valid options are 'grayscale', 'rgb' and 'hsv'
color_mode = grayscale
; download this file directly from NIH dropbox
data_entry_file = ./datasets/pxr_ds/image_data_entry.csv
; class names, you should not modify this
class_names = Intertrochanteric_FX,Femoral_Neck_FX,Normal
; use the following model (currently support densenet121)
nn_model = densenet121
[IMAGE-PREPROCESSING]
; normalize
normalize_by_mean_var = true
; color_mode=grayscale
normalize_mean = 0.0
; color_mode=rgb, hsv
normalize_mean_chan1 = 0.0
normalize_mean_chan2 = 0.0
normalize_mean_chan3 = 0.0
; color_mode=grayscale
normalize_stdev = 1.0
; color_mode=rgb, hsv
normalize_stdev_chan1 = 1.0
normalize_stdev_chan2 = 1.0
normalize_stdev_chan3 = 1.0
; samplewise zero-mean, variance normalization. DO NOT set true in conjection with normalize_by_mean_var=true
normalize_samplewise = false
[IMAGE-AUGMENATION]
; Image augmentation
; augmented images will be saved into
aug_verification_path = ./augmentation/verification
; enable augmentation in training set
train_augmentation = true
; enable augmentation in dev set
dev_augmentation = false
; enable augmentation in testing set
test_augmentation = false
random_horz_flip = true
random_vert_flip = true
[TRAIN]
; file path of imagenet pretrained weights, loaded at base_model initialization step
use_base_model_weights = true
base_model_weights_file = ./densenet121_grayscale.h5
; Resplit dataset (implies starting over)
force_resplit = false
; Print training progress
progress_verbosity = 1
; if true, load trained model weights saved in output_dir
; this is typically used for resuming your previous training tasks
; so the use_split_dataset will be automatically set to false
; also, make sure you use the reasonable initial_learning_rate
use_trained_model_weights = true
; if true, use best weights, else use last weights
use_best_weights = false
; note that the best weighting will be saved as best_weights.h5
output_weights_name = weights.h5
; basic training parameters
epochs = 10
batch_size = 32
initial_learning_rate = 0.001
; steps per epoch for training
; auto or int
; if auto is set, (total samples / batch_size) is used by default.
train_steps = auto
; steps per epoch for validation
; auto or int
; if auto is set, (total samples / batch_size) is used by default.
validation_steps = auto
; patience parameter used for ReduceLROnPlateau callback
; If val_loss doesn't decrease for x epochs, learning rate will be reduced by factor of 10.
patience_reduce_lr = 2
; this variable controlls the class_weight ratio between 0 and 1
; higher value means higher weighting of positive samples
positive_weights_multiply = 1
; if true, mean binary cross-entroy will be weighted by positive counts
; if false, just average the cross-entropy loss of each class
; in both cases, positive/negative sample ratio of each class is considered in the cross-entropy loss
use_class_balancing = true
; if true, use default split, otherwise create a new train/dev/test split
use_default_split = false
; random state used for splitting dataset into train/dev/test sets
split_dataset_random_state = 1
; print model summary
show_model_summary = false
[TEST]
; Print testing progress
progress_verbosity = 1
; Save grad-cam outputs
enable_grad_cam = true
; Output directory of gradcam images
grad_cam_outputdir = ./imgdir
batch_size = 32
test_generator_random_state = 1
; if true, use best_weights.h5, else use weights.h5
use_best_weights = true
- More baseline models
I would like to thank Pranav Rajpurkar (Stanford ML group) and Xinyu Weng (北京大學) for sharing their experiences on this task. Also I would like to thank Felix Yu for providing DenseNet-Keras source code.
Bruce Chou (brucechou1983@gmail.com) Michael (Tao-Yi) Lee (tylee@ieee.org)
Copyright 2018 (C) Bruce Chou, Michael (Tao-Yi) Lee
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.