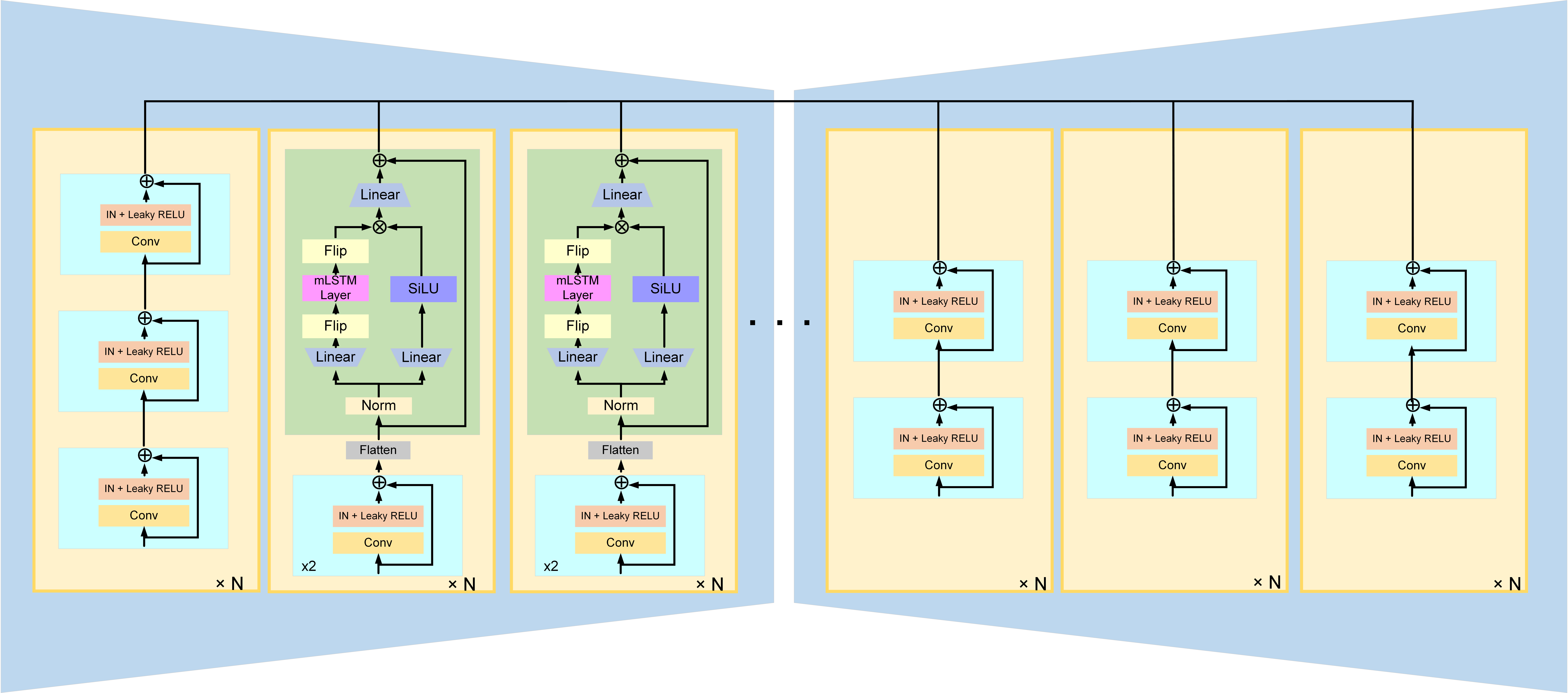
xLSTM-UNet can be an Effective 2D & 3D Medical Image Segmentation Backbone with Vision-LSTM (ViL) better than its Mamba Counterpart
Tianrun Chen, Chaotao Ding, Lanyun Zhu, Tao Xu, Deyi Ji, Ying Zang, Zejian Li
KOKONI, Moxin Technology (Huzhou) Co., LTD , Zhejiang University, Singapore University of Technology and Design, Huzhou University, University of Science and Technology of China.
Requirements: Ubuntu 20.04, CUDA 11.8
- Create a virtual environment:
conda create -n uxlstm python=3.10 -yandconda activate uxlstm - Install Pytorch 2.0.1:
pip install torch==2.0.1 torchvision==0.15.2 --index-url https://download.pytorch.org/whl/cu118 - Download code:
git clone https://github.com/tianrun-chen/xLSTM-UNet-PyTorch.git cd xLSTM-UNet-PyTorch/UxLSTMand runpip install -e .
The dataset used in this project is derived from the following research paper:
Jun Ma, Feifei Li, Bo Wang. "U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation." arXiv preprint arXiv:2401.04722, 2024.
Download dataset here and put them into the data folder. U-xLSTM is built on the popular nnU-Net framework.
Our data processing approach strictly follows the methods outlined in the U-Mamba study. This includes steps such as data normalization, augmentation techniques, and segmentation algorithms detailed in their publication.
nnUNetv2_plan_and_preprocess -d DATASET_ID --verify_dataset_integritynnUNetv2_train DATASET_ID {dataset_type} {exp_name} -tr {trainer_type} -lr {learning_rate} -bs {batch_size}-
{dataset_type}: Specifies the type of dataset to be used. There are two options:2d: For datasets that are 2-dimensional.3d_fullres: For full-resolution 3-dimensional datasets.
-
{exp_name}: Defines the name or identifier for the experiment. It can be:all: Use this option to include all available configurations.- An integer: Specify a particular configuration number to use a specific setup.
-
{trainer_type}: Indicates the type of trainer to use for the training process. Options include:nnUNetTrainerUxLSTMBot: A trainer tailored for the UxLSTMBot model.nnUNetTrainerUxLSTMEnc: A trainer designed for the UxLSTMEnc model architecture.
-
{learning_rate}: Specifies the learning rate for the training process. This should be a floating-point number. -
{batch_size}: Defines the number of samples to process before the model's internal parameters are updated.
- Train 2D
U-xLSTM_Botmodel
nnUNetv2_train DATASET_ID 2d all -tr nnUNetTrainerUxLSTMBot -lr {learning_rate} -bs {batch_size}- Train 2D
U-xLSTM_Encmodel
nnUNetv2_train DATASET_ID 2d all -tr nnUNetTrainerUxLSTMEnc -lr {learning_rate} -bs {batch_size}- Train 3D
U-xLSTM_Botmodel
nnUNetv2_train DATASET_ID 3d_fullres all -tr nnUNetTrainerUxLSTMBot -lr {learning_rate} -bs {batch_size}- Train 3D
U-xLSTM_Encmodel
nnUNetv2_train DATASET_ID 3d_fullres all -tr nnUNetTrainerUxLSTMEnc -lr {learning_rate} -bs {batch_size}- Predict testing cases with
U-xLSTM_Botmodel
nnUNetv2_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -d DATASET_ID -c DATASET_TYPE -f all -tr nnUNetTrainerUxLSTMBot --disable_tta- Predict testing cases with
U-xLSTM_Encmodel
nnUNetv2_predict -i INPUT_FOLDER -o OUTPUT_FOLDER -d DATASET_ID -c DATASET_TYPE -f all -tr nnUNetTrainerUxLSTMEnc --disable_tta
DATASET_TYPEcan be2dand3d_fullresfor 2D and 3D models, respectively.
- We provide pretrained models that you can download from this link. Please ensure you save them in the
pretrained_modelfolder. Then, run process_weight.py to verify that the weights are correctly placed in the appropriate file path
python process_weight.pyWe also provide additional shell scripts for evaluating our provided pretrained model.
bash metric_bot.sh
bash metric_enc.shIf you wish to evaluate your own trained model, be sure to change the -f parameter to the {exp_name} used during training. This could be 'all' or a specific integer identifier.
Please cite this work if you find it inspiring or helpful!
@misc{chen2024xlstmuneteffective2d,
title={xLSTM-UNet can be an Effective 2D \& 3D Medical Image Segmentation Backbone with Vision-LSTM (ViL) better than its Mamba Counterpart},
author={Tianrun Chen and Chaotao Ding and Lanyun Zhu and Tao Xu and Deyi Ji and Ying Zang and Zejian Li},
year={2024},
eprint={2407.01530},
archivePrefix={arXiv},
primaryClass={eess.IV},
url={https://arxiv.org/abs/2407.01530},
}
You are also welcomed to check our Segment Anything Adapter (SAM-Adapter)
@misc{chen2023sam,
title={SAM Fails to Segment Anything? -- SAM-Adapter: Adapting SAM in Underperformed Scenes: Camouflage, Shadow, and More},
author={Tianrun Chen and Lanyun Zhu and Chaotao Ding and Runlong Cao and Shangzhan Zhang and Yan Wang and Zejian Li and Lingyun Sun and Papa Mao and Ying Zang},
year={2023},
eprint={2304.09148},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
The code is based on Jun Ma, Feifei Li, Bo Wang. "U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation." arXiv preprint arXiv:2401.04722, 2024.