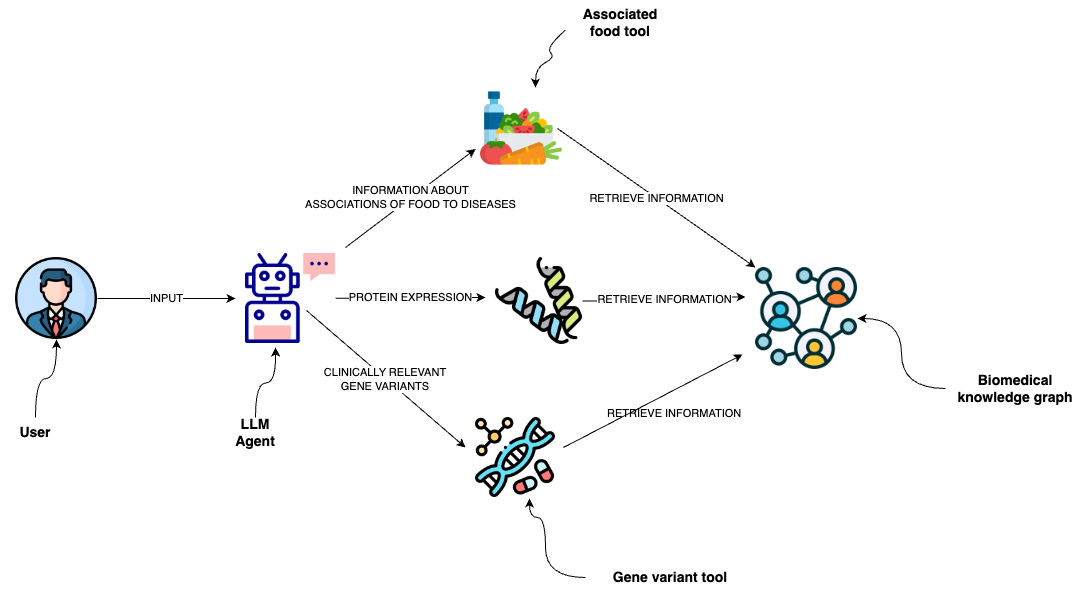
This project is designed for a simple LLM agent with tools on a clinical knowledge graph in Neo4j.
-
Download and restore the Clinical database dump.
-
Set environment variables in
.env. -
Run the following command:
docker compose up
- What are associated foods with cancer?
- What are known clinically relevant gene variants for gene CLN3?
- In which tissues are expressed proteins associated with prostate carcinoma?
- Better entity mapping from input to database (Using a combination of keyword search and/or biomedical text embeddings)
- More tools
Contributions are welcomed