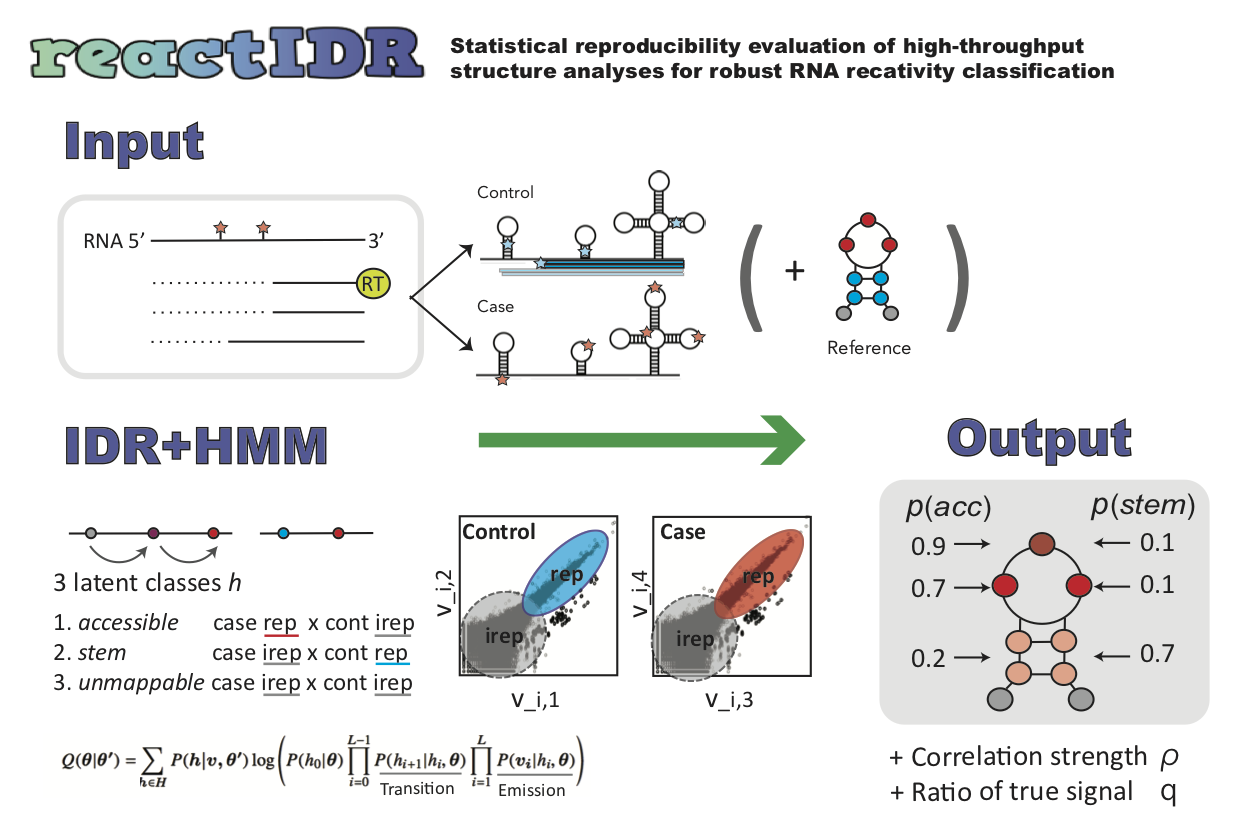
reactIDR: evaluation of the statistical reproducibility of high-throughput structural analyses towards a robust RNA structure prediction
- Published in BMC Bioinformatics
-
Input read count data
- PARS
- SHAPE-Seq
- icSHAPE
- DMS-Seq (assumed to be enriched only at A or C)
-
Output
- posterior probability of being loop (enriched in case) or stem (enriched in control)
-
Algorithm
- IDR + hidden Markov Model
- python3
- numpy
- scikit-learn
Other packages are required for visualization process as follows:
- pandas
- seaborn
git clone https://github.com/carushi/reactIDR
cd reactIDR/
python setup.py build_ext -b reactIDR/ # cython build
cd example && bash training.sh # Run test
Please visit our wiki for further info.
- read_collapse.py
- collapse PCR duplicates and trim barcode
- assume gawk
- read_truncate.py
- extract consistent paired end reads
- bed_to_pars_format.py
- write PARS-formatted 5' end coverage data based on gtf and gff annotation or sequence location
- format: NAME 0;1;2;3;.....
- tab_to_csv.py
- use to append raw count (read count, coverage, ...) to the output csv file
-
R. Kawaguchi, H. Kiryu, J. Iwakiri and J. Sese. "reactIDR: evaluation of the statistical reproducibility of high-throughput structural analyses towards a robust RNA structure prediction" BMC Bioinformatics 20 (Suppl 3) :130 (2019)" ー Selected for APBC '19 proceedings
-
- Convert bam to read count data
- Find scripts and how to use at https://github.com/carushi/RT_end_counter
-
IDR
- Li, Qunhua, et al. "Measuring reproducibility of high-throughput experiments", The annals of applied statistics, 2011.
- IDR in Python
- IDR in R
- apply to MaP analyses