-
Notifications
You must be signed in to change notification settings - Fork 8
Loading and editing networks
- Import networks from your file
- Import networks from databases
- Create your own network
- Search and filter your network
You can upload your own network from a xlsx, plain text .txt or a network sif file. Just go to File > Import > Network from SIF / XLSX / TXT. It is very easy, just browse your file and click ok.
####SIF FILE
If your network is already in sif format, use this option. See the link for more information about the SIF format.
- Browse your file. A preview of your data will be displayed in the table.
- Click ok.
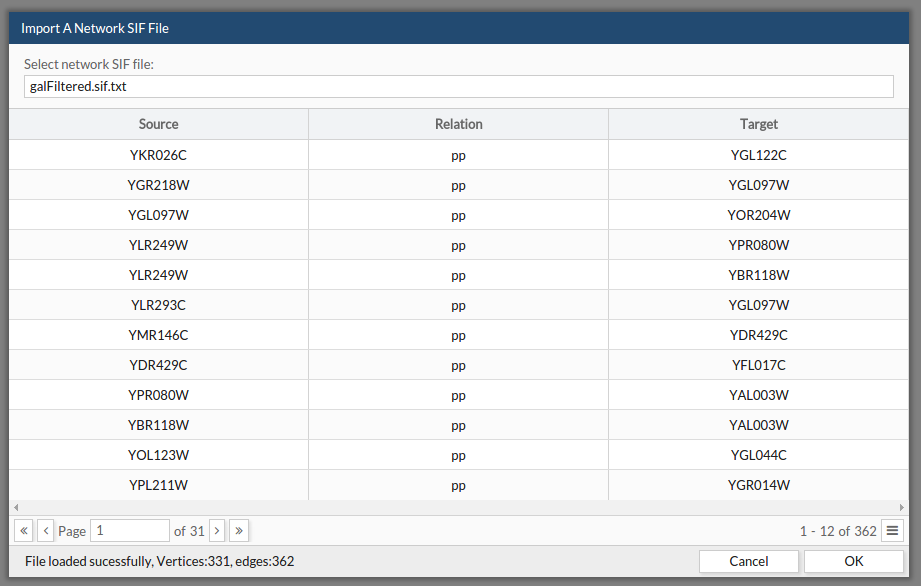
See the example file: example sif. This example can be found as the first example in the example section at the menu bar.
####XLSX FILE
Also, you can upload networks directly from a xlsx file:
- Select the file from your computer.
- Indicate in which sheet is your network.
- Indicate which column contains the node source, which one contains the edge type (if any) and the node target. Once you have told CellMaps all these instructions, a preview of your data will be displayed in the table.
- Click ok.
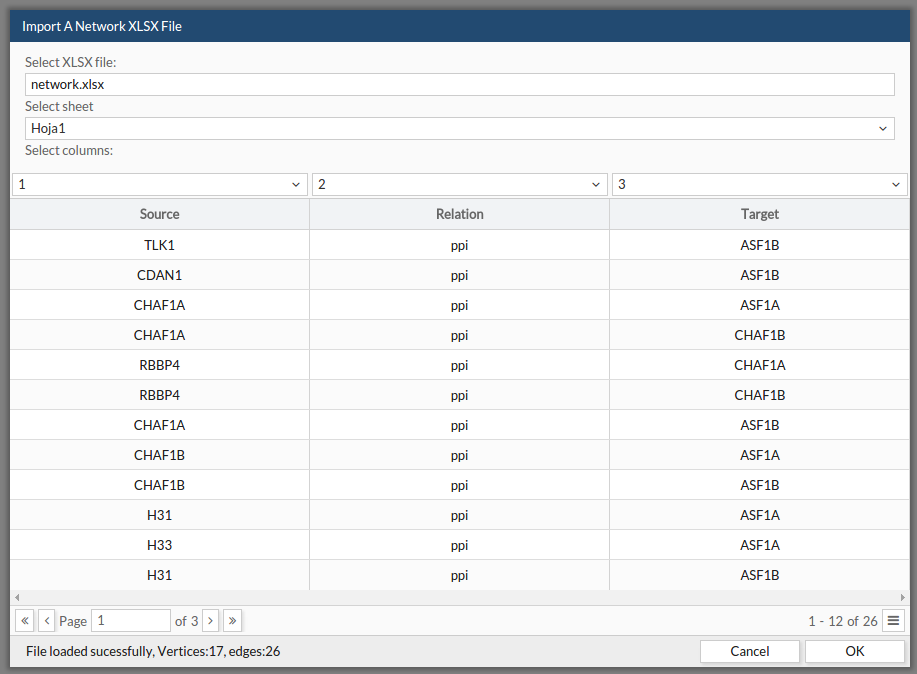
See the example file: example xlsx. This example can be found as the first example in the example section at the menu bar.
####TXT FILE
Finally, you can upload networks directly from a txt file:
- Select the file from your computer.
- Indicate which separator are you using to separate the columns/fields.
- Indicate which column contains the node source, which one contains the edge type (if any) and the node target. Once you have told CellMaps all these instructions, a preview of your data will be displayed in the table.
- Click ok.
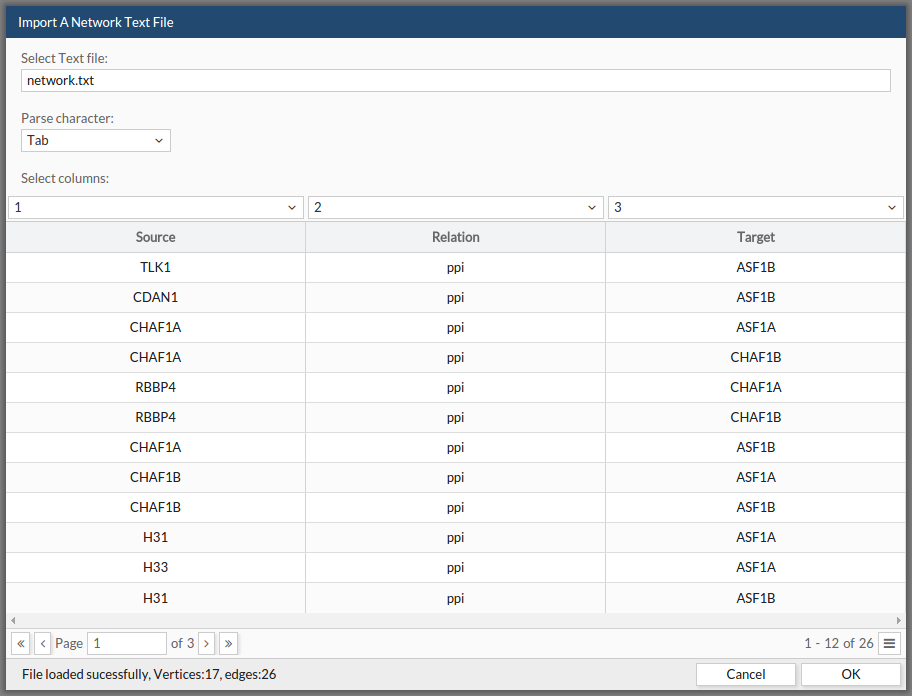
See the example file: example txt This example can be found as the first example in the example section at the menu bar.
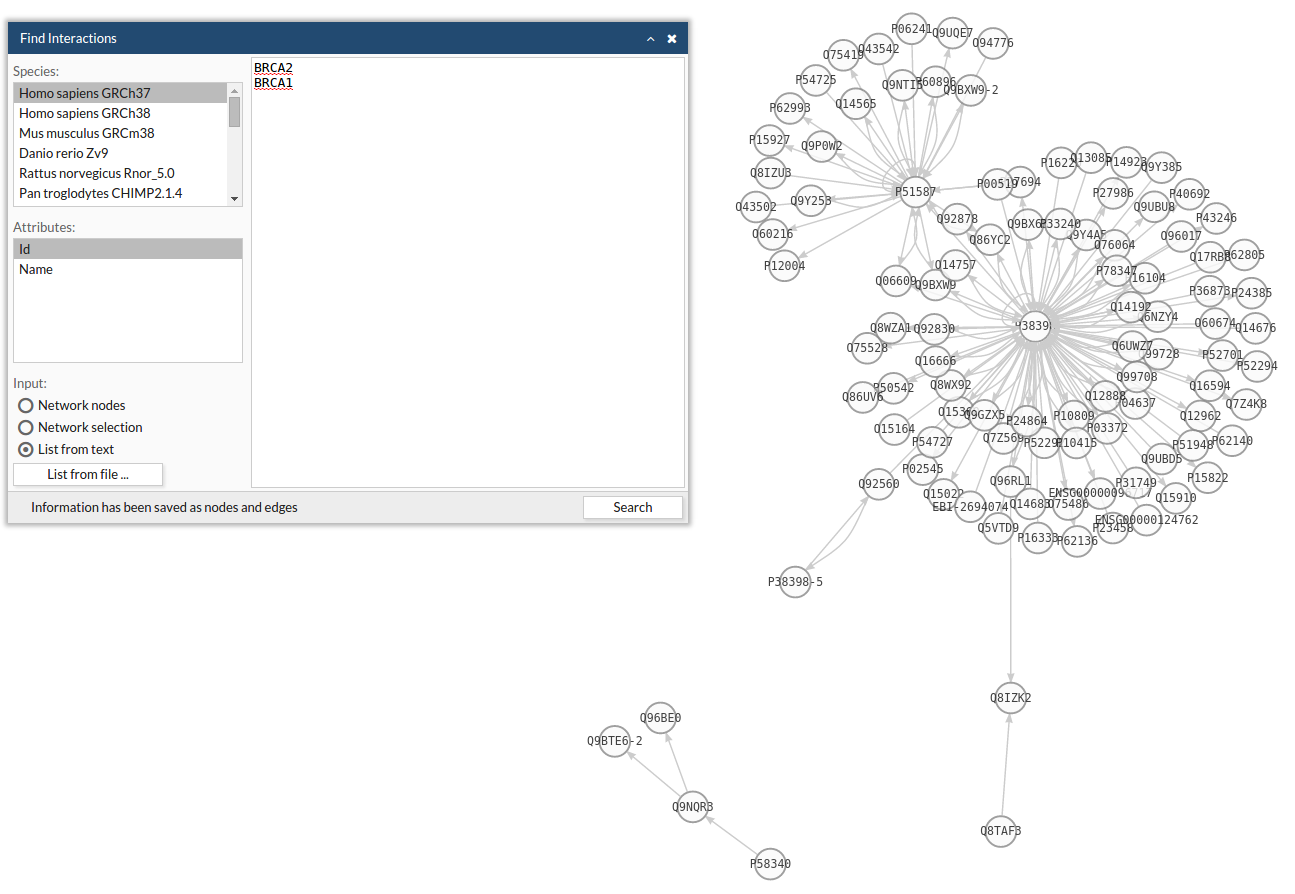
Use a network from an external database by going to Network > Import Network > IntAct. A popup window will appear.
- Select the specie you are working with.
- Introduce the genes for which you want to get the interactions. Here you can either:
- Use all nodes from the network you are working with
- Use only the selected nodes from the network you are working with
- Introduce a list of gene/proteins directly in the text box.
- Upload a list of gene/proteins from file.
- Click Enter and the network will appear in the viewer window.
At the toolbar there are the editor buttons that enables you to build and modify networks interactively by adding and removing nodes and edges onto the viewer window.
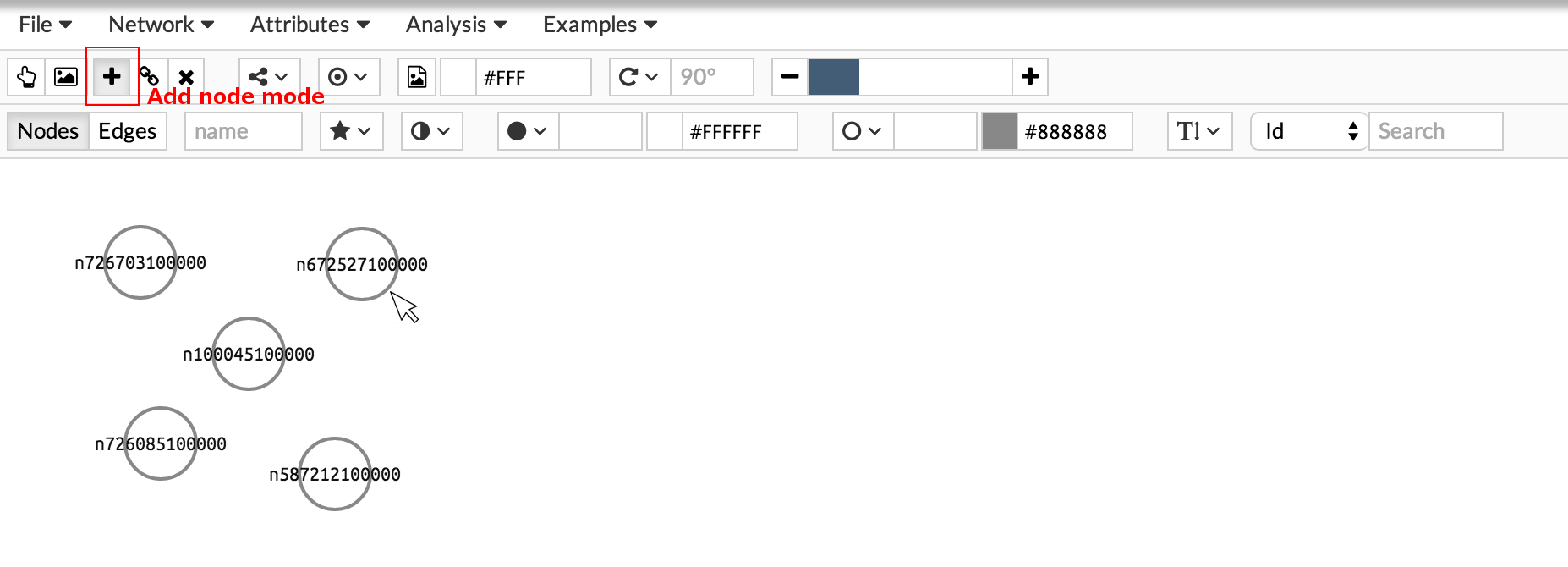
Start adding nodes by activating the add node button. Once activated, click on the viewer window to add the node.
Then, you can add an edge between two nodes by activating the join nodes button. To draw the edge, first click on the source node. Then, move the cursor to the target node (a line will follow the cursor) and click again on to create the connection.
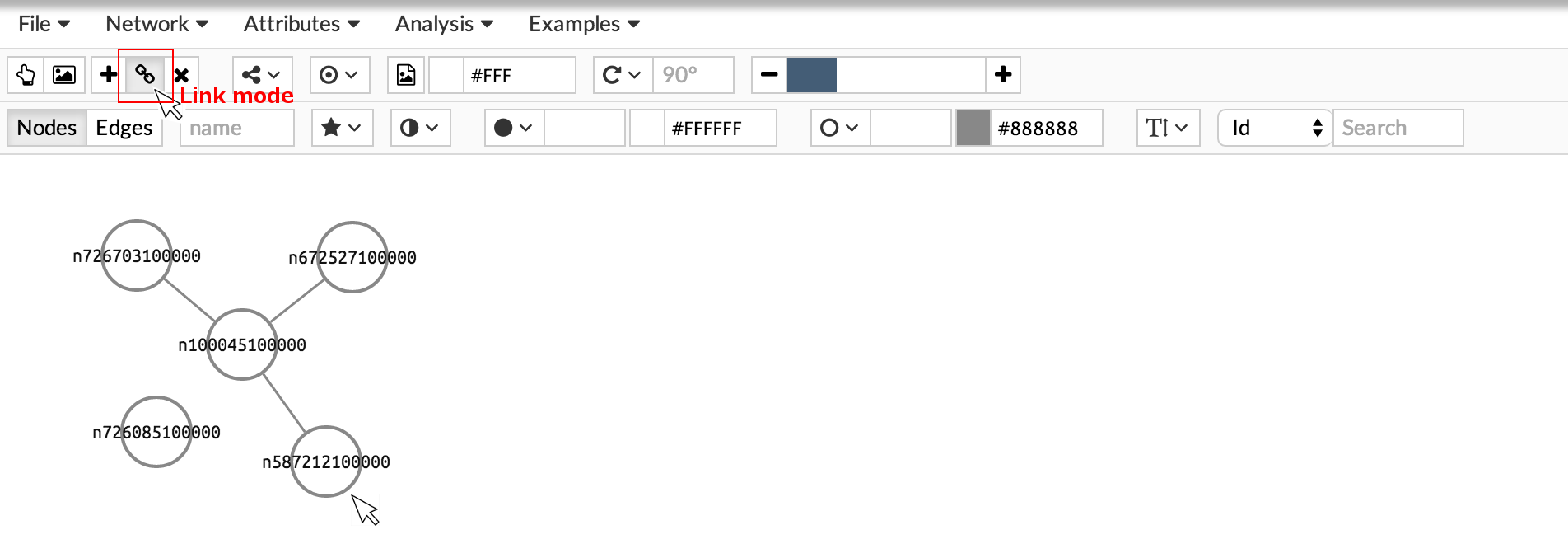
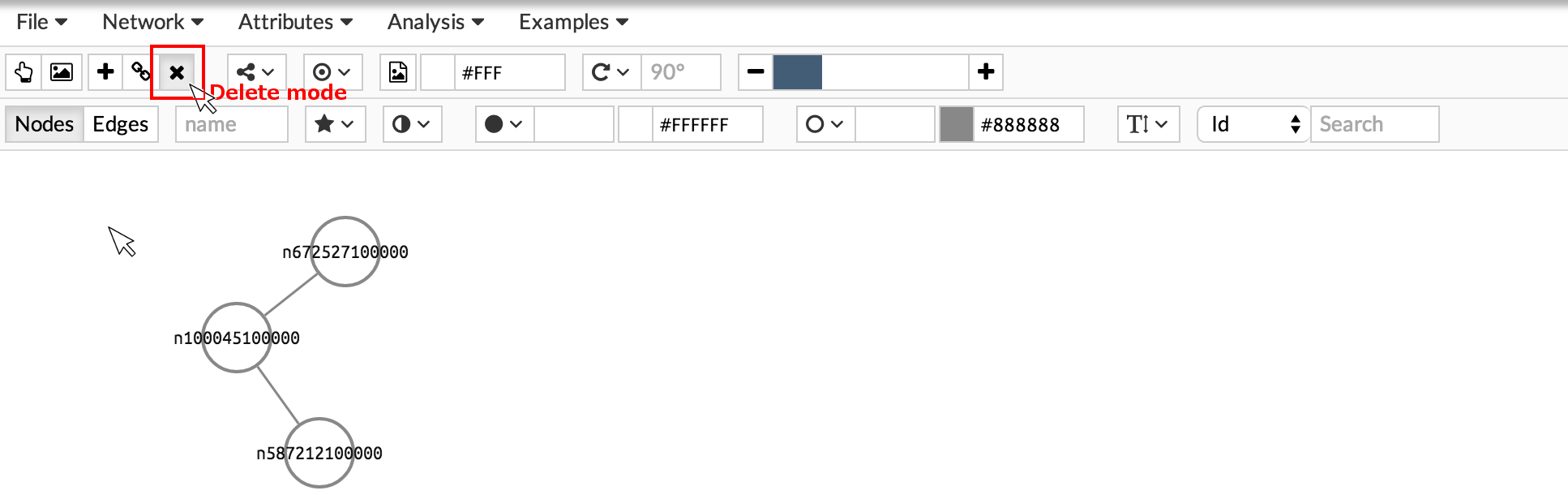
Remove nodes and edges by activating the delete button and clicking on the element you want to remove.
I you have integrated node or edge attributes (see here ), then you can use these to explore and filter your network. Go to Network > Search > Nodes/edges by attributes. A box will appear when you can create as many queries as you need.
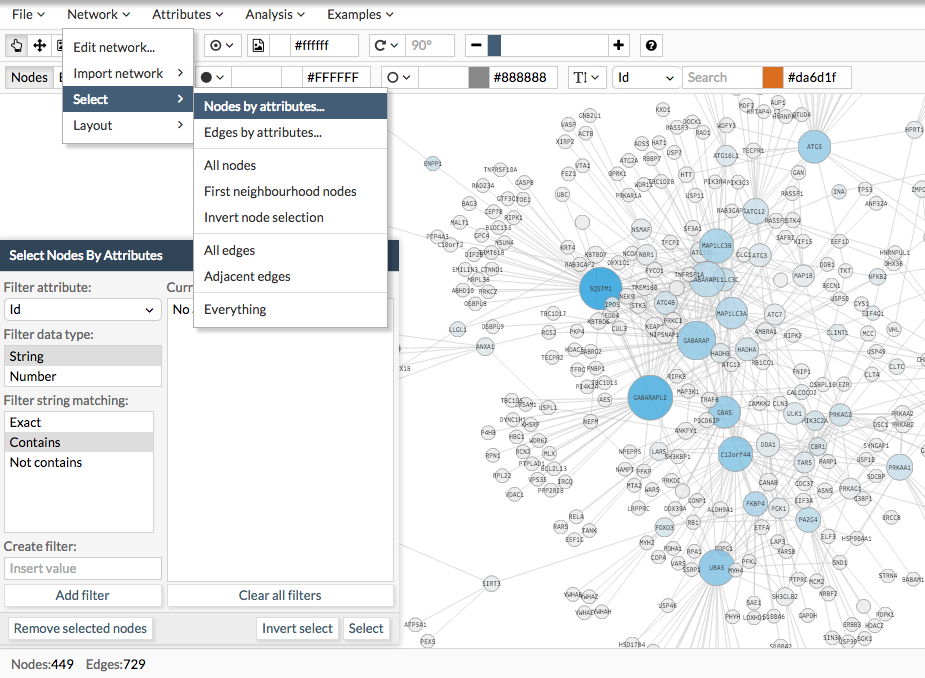
In the image below describes the how the filter box works:

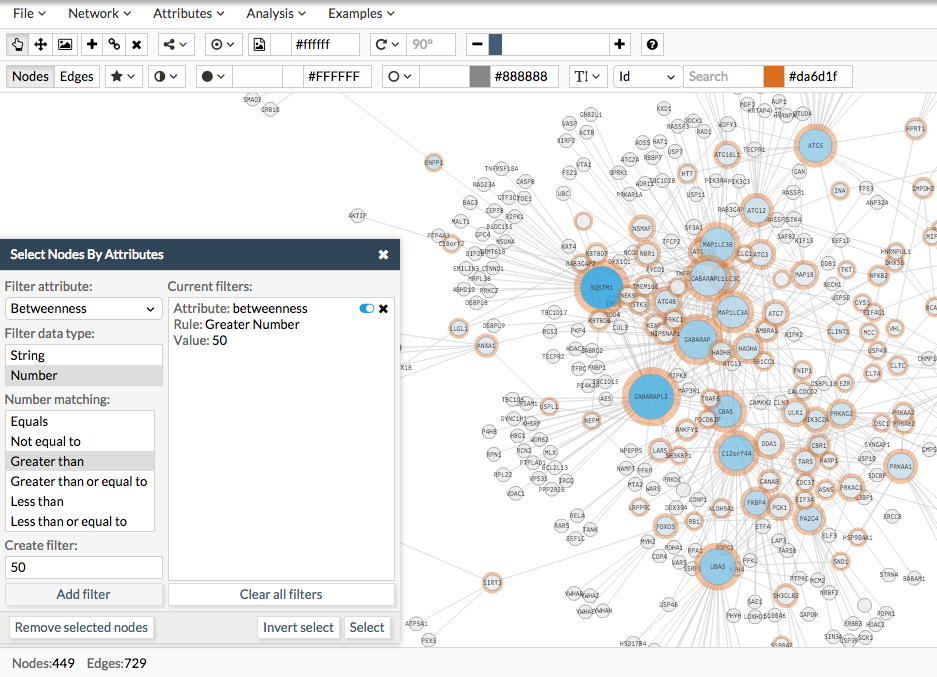
For example, in the following image we describe how to select nodes with a betweenness greater than 50. Configure your query, click add filter and then click on select (or in the inverted select). Those nodes matching the criteria will be highlighted. If you want to remove them, then choose remove. If you want to change its visual style, you can use the toolbar. Also, you can create a new attribute and add a specific values for the selected nodes by going to the menu Attributes>Edit node attributes.
Note that when you remove nodes, these will be completely removed from your actual session and can not be recovered. We recommend you to save a session before starting the filters so you always will be able to come back if you change your mind.