-
Notifications
You must be signed in to change notification settings - Fork 8
Saving sessions and exporting data
This section describes how you can save a session, open a previous session, and manage your saved sessions.
After working with CellMaps users can download the session in JSON format just selecting Save session from the File menu. Users will be asked to give a name to the file and a location. Sessions will store any network, attributes, background images and configurations (colors, mappings, etc.) created during the session. Sessions can be saved without the need to be logged in.
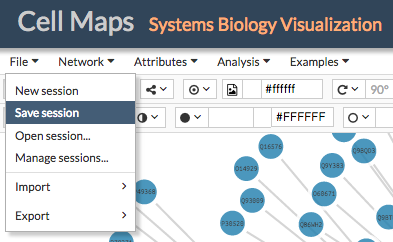
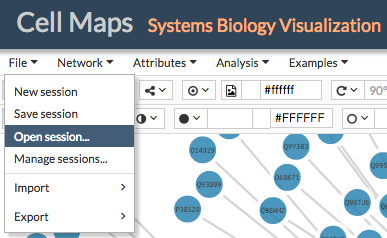
To load a previous session go to File > Open session... and select a previously stored session in JSON format.
Logged users will be able to store CellMaps sessions in our servers without needing to download and upload them. To store a session in our servers just go to File > Manage sessions.... Give a name to the session and click Add session. A new entry will appear in the list of Saved sessions.
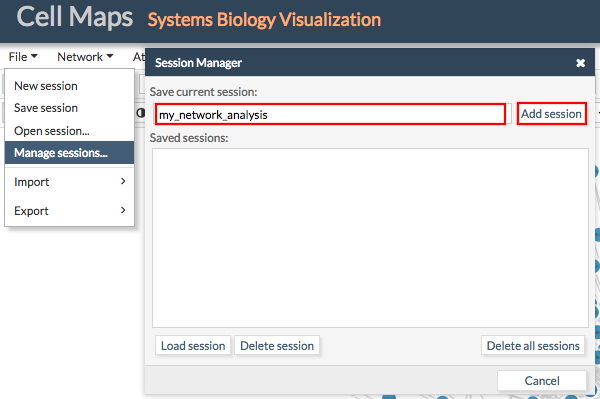
To load an old session just select the desired session from the list of Saved sessions and click Load session.
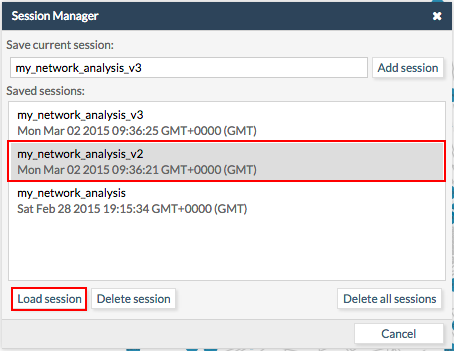
The same way users can delete sessions.
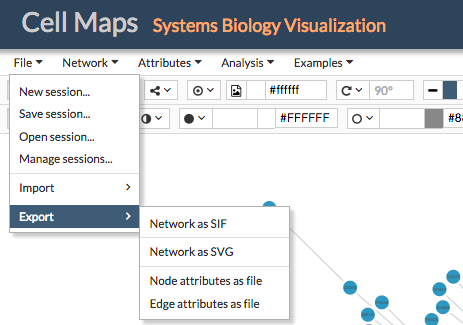
CellMaps allow the user to export network-related data in several formats. By going to File > Export, users can chose between the following formats:
- Network as SIF: A file containing the interactions of the current network in SIF format. This file does not include attributes and user configurations, to download this information go to Saving a session
- Network as SVG: An image of the current network viewer in SVG format
- Node attributes as file: A tabular file containing the uploaded and downloaded attributes for the nodes in the current network. The first column in the file indicates the node identifier and the rest of the columns correspond to the used node attributes.
- Edge attributes as file: A tabular file containing the uploaded and downloaded attributes for the edges (or connections) in the current network. The first column in the file indicates the edge identifier (generally, nodeA_interactionType_nodeB, e.g: BRCA2_pp_BCL2) and the rest of the columns correspond to the used edge attributes.