-
Notifications
You must be signed in to change notification settings - Fork 0
08. SNP and InDel detection
Sebastian Gregoricchio edited this page Oct 29, 2023
·
10 revisions
Calling of genetic variants requires a relatively high read coverage in order to be reliable. The amount of reads sequenced for ChIp-seq is usually not in the range required for the variant calling. However, ChIP-seq peaks display a local deep coverage of that specific genomic region. Hence, SPACCa will merge all the peaks individuated and perform a variant calling only in this regions for each sample by using the golden standard tool GATK (HaplotypeCaller). Then, variants are filtered and annotated by SnpEff & SnpSift based on the user-provided dbSNP database.
| Parameter | Description |
|---|---|
| call_variants |
True`False` to indicate whether to perform variant calling at peaks. |
| dbsnp_file | SNP database file (.dbsnp) for base recalibration required by GATK4. It could happen that your .bam files contain the 'chr' prefix in the chromosome names while your dbSNP file does not (or vice versa). This can be fixed in the .dbsnp file with the bcftools annotate --rename-chrs command. For Hg38, for instance, the dbSNP file can be downloaded from the broad institute cloud storage. Do not forget to download the INDEX as well! |
| DP_snp_threshold | Default: 20. Threshold for the hard-filtering of the SNP (single-nucleotide polymorphism) VCF table for sequencing depth (DP) by SnpSift Filter. |
| QUAL_snp_threshold | Default: 0. Threshold for the hard-filtering of the SNP (single-nucleotide polymorphism) VCF table for quality (QUAL) by SnpSift Filter. |
| DP_indel_threshold | Default: 20. Threshold for the hard-filtering of the InDel (insertion/delition) VCF table for sequencing depth (DP) by SnpSift Filter. |
| QUAL_indel_threshold | Default: 0. Threshold for the hard-filtering of the InDel (insertion/delition) VCF table for quality (QUAL) by SnpSift Filter. |
| SnpSift_vcf_fields_to_extract | Default: [ "CHROM", "POS", "ID", "REF", "ALT", "QUAL", "DP", "AF", "FILTER", "FORMAT", "GEN[*].GT", "GEN[*].AD", "GEN[*].AF" ]. A python list to indicate the fields to be extracted from the VCFs (both SNP and InDel) during the conversion to .txt file by SnpSift Extract Fields. |
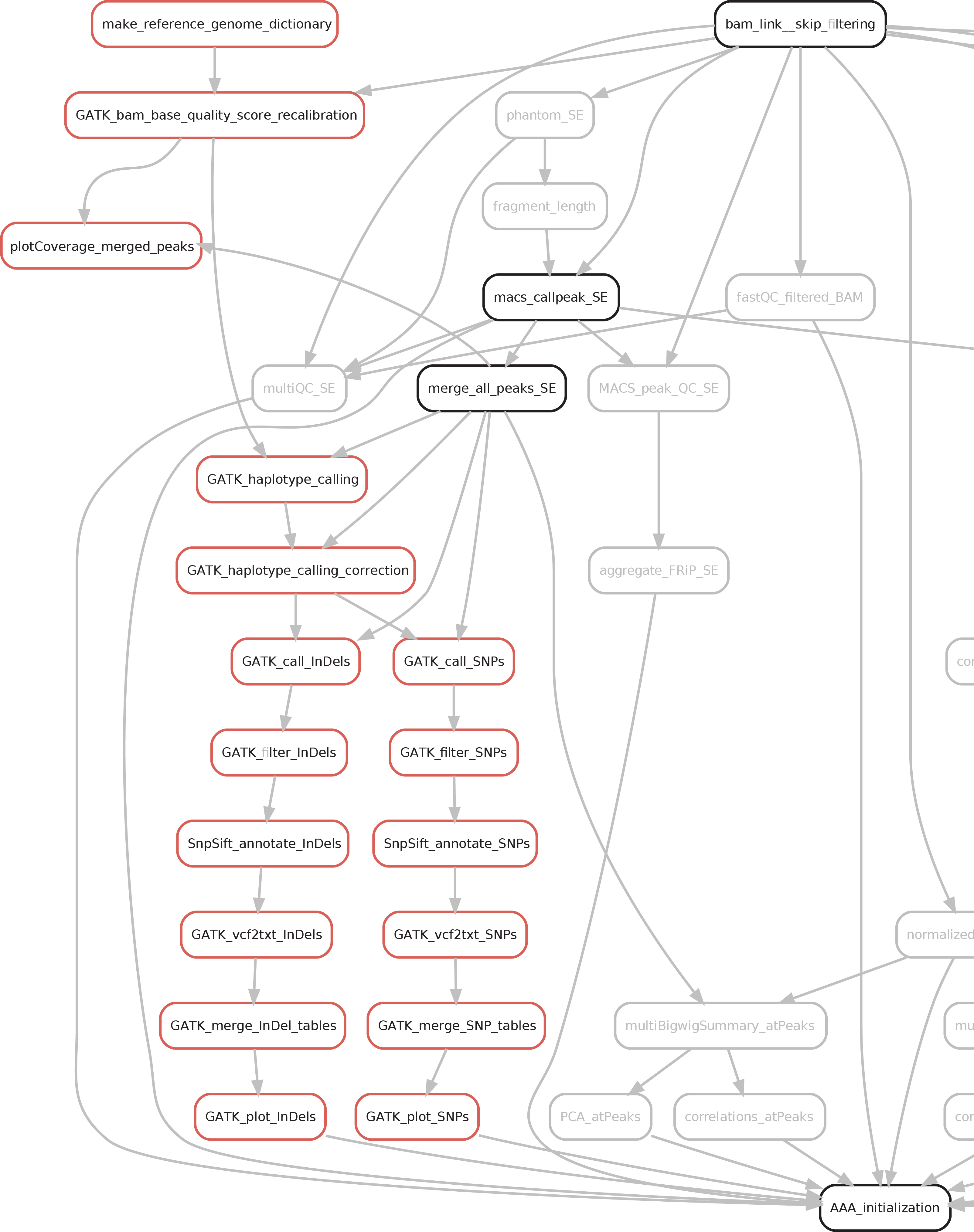
The results of the variant calling can be found in the 06a_Somatic_variants directory, which will contain:
- recalibrated_bams: bam files will undergo to Base Quality Score Recalibration (BSQR) and stored in this folder
- coverage_plots: for each sample there is a file with two plots. The first represents the frequencies of the found read coverage at the peaks, the second instead shows a distribution estimating the fraction of genome that has a specific sequencing depth 9for details see the plotCoverage deepTools page).
- VCF: in this folder are contained unfiltered variants (g.vcf files), and filtered .vcf and .txt tables of variants detected in each samples as well as a merged table with all the samples combined 9a column indicates the corresponding sample)
- SV_count_plots: two bar plots, one for SNP and one for InDel, depict the number of SNP/InDel found for each sample
output_folder
...
└── 06a_Somatic_variants
├── all_samples_peaks_concatenation_collapsed_sorted.bed
├── coverage_plots
│ └── sample_plotCoverage.pdf
├── recalibrated_bams
│ ├── bsqr_tables
│ │ └── sample_mapQ20_sorted_woMT_mdup_bsqr.table
│ ├── sample_mapQ20_sorted_woMT_mdup_bsqr.bam
│ └── sample_mapQ20_sorted_woMT_mdup_bsqr.bai
├── SV_count_plots
│ ├── all.samples_InDel_counts_plot.pdf
│ ├── all.samples_SNP_counts_plot.pdf
└── VCF
├── InDel
│ ├── all.samples_mdup_gatk-indel_filtered.DP20.QUAL0_annotated.txt
│ ├── sample_dedup_gatk-indel_filtered.DP20.QUAL20_annotated.txt
│ ├── sample_dedup_gatk-indel_filtered.DP20.QUAL20_annotated.vcf.gz
│ └── sample_dedup_gatk-indel_filtered.DP20.QUAL20_annotated.vcf.gz.tbi
└── SNP
├── all.samples_mdup_gatk-snp_filtered.DP20.QUAL0_annotated.txt
├── sample_dedup_gatk-snp_filtered.DP20.QUAL20_annotated.txt
├── sample_dedup_gatk-snp_filtered.DP20.QUAL20_annotated.vcf.gz
└── sample_dedup_gatk-snp_filtered.DP20.QUAL20_annotated.vcf.gz.tbi
Contributors